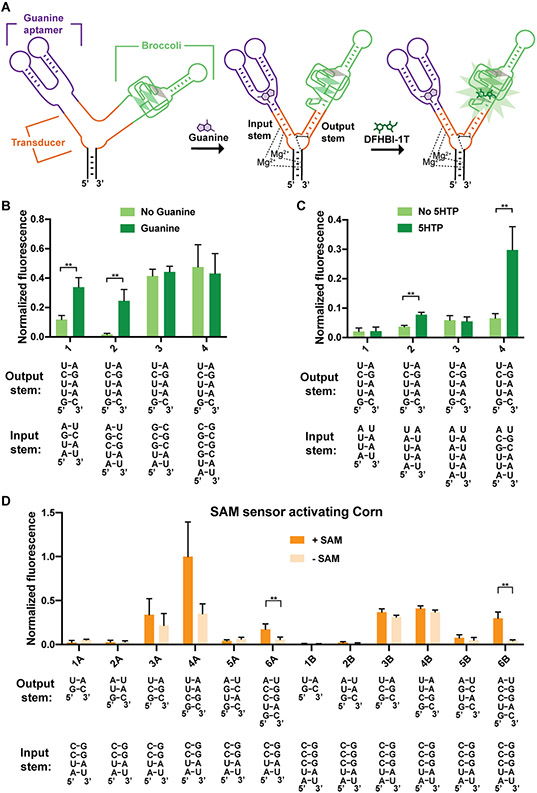
Figure 5. Optimizing F30-based sensors composed of different input and output aptamers.
(A) Schematic representation of an F30 three-way junction-based guanine sensor. Guanine binding to the guanine aptamer induces folding of the input stem, which facilitates the proper positioning of the U-rich core domain (shown are the H-bonds and the base stacking interactions of the core) and the output stem. The positioning of the U-rich core domain is expected to stabilize the output stem and promote the folding of Broccoli. (B) Optimization of F30-based guanine sensors. RNA (4 μM) was incubated with 40 μM DFHBI-1T with or without 0.2 mM guanine. Fluorescence of each sample was measured (Ex 460 nm, Em 505 nm) and normalized to Broccoli. The mean and SEM values are shown (n=3). **P = 0.005 (1), **P = 0.007 (2). (C) Optimization of input and output stems in F30-based 5HTP sensors. The mean and SEM values are shown (n=3). **P = 0.001 (2), ** P = 0.008 (4). (D) Optimization of F30-based SAM sensors with Corn as the output aptamer. RNA (0.5 μM) was incubated with 10 μM DFHO with or without 0.2 mM SAM. After 1 h, fluorescence was measured and normalized to the average fluorescence of combination 4A (the brightest combination tested). The mean and SEM values are shown (n=3). *P = 0.04 (6A), **P = 0.004 (6B).