Abstract
The proper DNA damage response (DDR) and repair are the central molecular mechanisms for the maintenance of cellular homeostasis and genomic integrity. The abnormality in this process is frequently observed in human cancers, and is an important contributing factor to cancer development. FBXW7 is an F-box protein serving as the substrate recognition component of SCF (SKP1-CUL1-F-box protein) E3 ubiquitin ligase. By selectively targeting many oncoproteins for proteasome-mediated degradation, FBXW7 acts as a typical tumor suppressor. Recent studies have demonstrated that FBXW7 also plays critical roles in the process of DDR and repair. In this review, we first briefly introduce the processes of protein ubiquitylation by SCFFBXW7 and DDR/repair, then provide an overview of the molecular characteristics of FBXW7. We next discuss how FBXW7 regulates the process of DDR and repair, and its translational implication. Finally, we propose few future perspectives to further elucidate the role of FBXW7 in regulation of a variety of biological processes and tumorigenesis, and to design a number of approaches for FBXW7 reactivation in a subset of human cancers for potential anticancer therapy.
Keywords: cancer, DDR, DNA repair, FBXW7, ubiquitylation
Introduction
Protein Ubiquitylation and SCF E3 Ligase With FBXW7 as a Substrate Receptor
Ubiquitylation is a typical post-translational modification, that couples with proteasome, designated as ubiquitin-proteasome system (UPS), as the key proteolytic mechanism in eukaryotes for timely degradation of cellular proteins (Hershko et al., 2000). In general, the UPS-mediated protein degradation includes two steps: (1) covalent attachment of the small peptide ubiquitin to a substrate, a process called ubiquitylation; (2) delivery of ubiquitylated substrates into 26S proteasome for degradation. Ubiquitylation is a well-defined three-step enzymatic cascade catalyzed sequentially by the ubiquitin-activating enzymes (E1s), ubiquitin-conjugating enzymes (E2s), and ubiquitin ligases (E3s) (Ciechanover, 1998). Crucially, E3s determine the substrate specificity through selectively recognizing and directly binding with substrate proteins doomed for ubiquitylation and subsequent degradation.
Among the estimated >600 human E3 ubiquitin ligases, SCF (SKP1-CUL1-F-box protein) is the best studied member of CRL (Cullin-RING-Ligase) family of E3 enzymes. The SCF is a multi-component E3, consisting of a scaffold cullin-1, an adaptor SKP1, a E2 binding RING-domain protein (RBX1/RBX2), and a substrate-receptor F-box protein (Zhao and Sun, 2013; Figure 1A). Although mammalian genome contains 69 F-box proteins (Jin et al., 2004), only three, namely FBXW7, β-TrCP, and SKP2 are well defined and characterized (Skaar et al., 2014). Among these three, FBXW7 is a typical tumor suppressor that promotes the ubiquitylation and degradation of many cellular oncoproteins, and is frequently mutated and inactivated in many human cancers (Welcker and Clurman, 2008; Wang et al., 2012; Figure 2).
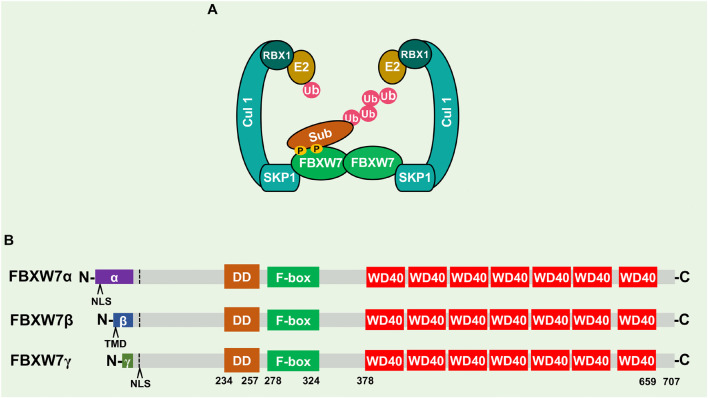
FIGURE 1.
Schematics of SCF, FBXW7, and FBXW7 isoforms. (A) SCFFBXW7 consists of a scaffold cullin-1 (CUL-1), an adaptor SKP1, a RING-domain protein (RBX1/RBX2), and a substrate-receptor F-box protein (FBXW7). Shown is SCFFBXW7 complex in FBXW7 dimerization format for ubiquitylation of a substrate. (B) Three FBXW7 isoforms (α, β, and γ) with domain alignment. NLS, nuclear localization signal; TMD, transmembrane domain; DD, dimerization domain.
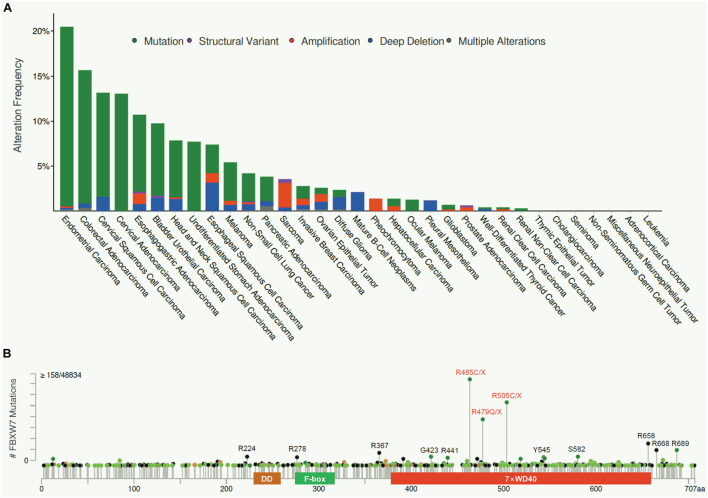
FIGURE 2.
FBXW7 mutation frequency in human cancers and their distributions. (A) FBXW7 mutation frequency in different types of human cancer, analyzed from cBioPortal Database (https://www.cbioportal.org/). (B) Distribution of FBXW7 mutations in the FBXW7 encoding region. Mutations detected in more than 15 cancer samples are annotated, and three most frequent mutation hotspots, R465, R479, and R505 are highlighted in red.
General Introduction of DNA Damage Response and Repair
Mammalian cells are constantly exposed to external and internal insults, such as ionizing radiation (IR), ultraviolet (UV) light, reactive oxygen species (ROS), and many cellular metabolites. These insults typically cause genomic DNA damage in the forms of single strand breaks (SSBs), double strand breaks (DSBs) or replication fork stagnation, and the other types. Among them, DSBs is the most toxic form of DNA damage (Finn et al., 2012).
Upon DSB, a typical DDR is triggered in a cell in an attempt to repair damaged DNA to maintain the genomic integrity. Specifically, three phosphoinositide 3-kinases: ATM (ataxia telangiectasia mutated), ATR (ATM-and RAD3-related), and DNA-PK (DNA-dependent protein kinase) are first recruited into the damage sites and activated via auto-phosphorylation. They then phosphorylate H2AX into γH2AX, which directly binds with MDC1 (mediator of DNA damage checkpoint 1), and recruits the MRN (MRE11-RAD50-NBS1) protein complex to accumulate within the damage sites. The MRN complex acts as an amplifier of DDR signals by enhancing the activity of ATM (Sancar et al., 2004), whereas γ-H2AX is a key mediator for recruitment and retention of high concentration of DNA damage repair enzymes, such as 53BP1, RAD51, and BRCA1 in the vicinity of damaged sites (Nakamura et al., 2010). Following the expansion of DDR signals, two DNA damage repair machineries, namely homologous recombination (HR) and non-homologous end joining (NHEJ), are triggered rapidly and executed to repair the damaged DNA. The failure in DDR and repair is the cause of genomic instability, leading to cell death (if the damage is severe), or various gene mutations to trigger tumorigenesis (Srivastava and Raghavan, 2015).
During the process of DNA damage response and repair, the chromatin and repairing factors are regulated by a spectrum of post-translational modifications including phosphorylation, acetylation, methylation, and ubiquitylation (Wurtele and Verreault, 2006; Van and Santos, 2018). In particular, FBXW7, a receptor protein of SCF E3 ubiquitin ligase, has been recently shown to play fundamental roles in DDR and repair, which is the focus of this review.
Molecular Characterization of FBXW7
Isoforms and Subcellular Localizations
The human FBXW7 gene is localized at chromosome 4q32, a region deleted in 30% of cancers, and encodes three different isoforms (FBXW7α, β, and γ), derived from alternative splicing of the same transcript (Davis et al., 2014). These three isoforms share 10 common exons, encoding three conserved functional domains: (1) the DD dimerization domain, (2) F-box domain to recruit other SCF components, essential for its E3 ligase activity, and (3) substrate recognizing WD40 domain (Hao et al., 2007; Figure 1B). Three isoforms vary at the N-terminus and have different subcellular locations with FBXW7α in the nucleoplasm, FBXW7β in the cytoplasm, and FBXW7γ in the nucleolus (Davis et al., 2014). FBXW7α is functionally the most dominant isoform, which is ubiquitously expressed in most human tissues; FBXW7β expression is mainly found in brain and testis, whereas FBXW7γ is poorly understood and expressed mainly in muscles (Spruck et al., 2002; Matsumoto et al., 2006).
Dimerization
Dimerization is a common phenomenon and key regulatory modality for FBXW7 (Tang et al., 2007; Welcker and Clurman, 2007; Welcker et al., 2013), as well as for other F-box proteins, such as β-TrCP (Suzuki et al., 2000) and SKP2 (Inuzuka et al., 2012). The FBXW7 dimerization is mediated by the DD domain which enhances its catalytic efficiency for substrate degradation with two possible underlying mechanisms (Tang et al., 2007; Welcker et al., 2013): (1) Dimerization enhances the binding affinity between FBXW7 and substrates. Specifically, the dimer form of SCFFBXW7 provides spatial variability for accommodating diverse acceptor lysine geometries in both substrates and ubiquitin chain; and (2) The dimer-orthologs may provide suboptimal and independent recognition sites for substrates, serving as a complementary “buffer” against deleterious mutations in the WD40 domain (Welcker et al., 2013). Furthermore, under overexpressed conditions, FBXW7 could form the stable dimeric form by preventing autoubiquitylation of the monomeric form (Min et al., 2012; Lan et al., 2019). However, a contradictory study showed that while endogenous monomers and dimers are equally stable, the exogenous FBXW7 monomers appears to be more stable than that of dimers (Welcker et al., 2013). Exact reason for this discrepancy is unclear. The authors used wild-type FBXW7 monomer coupled with ubiquitylation-dead FBXW7ΔF monomer and proposed that trans-autoubiquitylation may be a major destabilization mechanism. Another possibility is that FBXW7 overexpression may trigger a limiting factor, such as a deubiquitylating enzyme, to block FBXW7 degradation (Welcker et al., 2013).
Recently, we and the others found that FBXW7 dimerization was regulated by several FBXW7-interacting proteins. For example, the prolyl isomerase PIN1 interacts with FBXW7 to prevent its dimerization in a phosphorylation-dependent manner (Min et al., 2012), whereas LSD1 directly binds to FBXW7 to disrupt FBXW7 dimerization, leading to its self-ubiquitylation (Lan et al., 2019).
Phosphorylation
FBXW7 was also regulated by phosphorylation which also affects its stability. One study showed that extracellular signal-regulated kinase (ERK) directly interacted with and phosphorylated FBXW7 at Thr205, which promoted FBXW7 ubiquitylation in a PIN-1 dependent manner in pancreatic cancer cells, although exactly how ERK-mediated FBXW7 phosphorylation triggers FBXW7 ubiquitylation remains elusive (Ji et al., 2015). Nevertheless, the study revealed a new mechanism by which the Kras-MAPK signal promotes pancreatic tumorigenesis via promoting degradation of tumor suppressor FBXW7. Another study showed that PLK1 phosphorylates FBXW7 at Ser58 and Thr284 to promote FBXW7 self-ubiquitylation, leading to stabilization of FBXW7 substrate N-MYC, which in turn transactivates PLK1, thus establishing a positive feed-forward loop that enhance MYC-regulated oncogenic programs (Xiao et al., 2016). On the contrary, two studies showed that FBXW7 phosphorylation at Ser227 by either serum and glucocorticoid-regulated kinase 1 (SGK1) or phosphoinositide 3-kinase (PI3K), respectively, switched the catalytic activity of FBXW7 toward its substrates instead of targeting itself for self-ubiquitylation (Mo et al., 2011; Schulein et al., 2011). Thus, FBXW7 phosphorylation appears to play critical role in FBXW7 stability in the manner dependent of the kinases and their phosphorylation sites. In addition, protein kinase (PK) C mediated FBXW7 phosphorylation at Ser8/Ser10 was reported to be involved in the nuclear localization of FBXW7α (Durgan and Parker, 2010).
Mutations in Human Cancers
Consistent with its role as a tumor suppressor, FBXW7 is the most frequently mutated gene among all the genes encoding F-box proteins in human cancers. We performed the meta-analyses of the cBioPortal Database1 and found an overall FBXW7 somatic mutation rate of 3.23% in human cancers (1,497 cases out of 46,305 tested), though different cancer types exhibit different mutational spectra (Figure 2A). The cancer with the highest mutational frequency is endometrial cancer (20.5%), followed by colorectal cancer (15.7%), cervical cancer (13.6), and esophagogastric adenocarcinoma (10.7%). Significantly, most of the FBXW7 mutations are single nucleotide change, resulting in single amino acid substitutions, within the WD40 domains responsible for substrate binding. As such, the mutations of these key residues often disrupt FBXW7 binding with its oncogenic substrates. Figure 2B showed three mutation hotspots R465, R479, and R505 (Welcker and Clurman, 2008; Davis et al., 2014), representing as much as 27.3% of cases (408/1497) found in all FBXW7 mutations, along with other mutation sites detected in more than 15 cases in all human cancers, including R224, R278, R367, G423, R441, Y545, S582, R658, R668, and R689.
Known FBXW7 Substrates
FBXW7 is well-known for its tumor suppressor function against cancer development by targeting a variety of oncoproteins for proteasomal degradation. A majority of FBXW7 substrates identified and characterized to date were summarized in Table 1. Almost all substrates contain a evolutionarily conserved phosphorylation motif, designated as CDC4 phospho-degron (CPD), of which substrate phosphorylation is a prerequisite event for FBXW7 binding and subsequent ubiquitylation (Nash et al., 2001). Among all FBXW7 substrates, most of them are well-known oncoproteins, which play the key roles in regulation of cell growth, apoptosis, differentiation and cell migration among the others (Wang et al., 2012). Some of these oncogenic substrates, such as c-MYC, NOTCH 1, MCL-1, Cyclin E, and c-JUN, likely play a driver role in FBXW7-associated cancers (Fryer et al., 2004; Wei et al., 2005; Welcker and Clurman, 2008; Davis et al., 2014). Taking c-MYC as an example, the studies using both in vitro and in vivo models showed that c-MYC is a classic substrate of FBXW7. Specifically, FBXW7-mediated c-MYC degradation relies on prior CPD phosphorylation of C-MYC at Thr58 and Ser62 by GSK3 and MAPK, respectively (Welcker et al., 2004; Yada et al., 2004). It is, therefore, not surprising that the point mutations on Thr58 and Ser62 were found on c-Myc in a variety of human cancers, thus avoiding FBXW7 degradation and being selected with growth advantage (Bahram et al., 2000). Moreover, in several fbxw7 KO mouse models, c-Myc was remarkably accumulated to accelerate tumorigenesis and promote tumor growth (Yada et al., 2004; Onoyama et al., 2007).
TABLE 1.
Summary of FBXW7 substrates.
| Substrates | Functions/Pathways | Kinase(s) | References |
| Aurora A | Protein kinase | GSK3 | Kwon et al., 2012 |
| Aurora B | Protein kinase | – | Teng et al., 2012 |
| BLM | DNA helicase | GSK3, CDK2 | Kharat et al., 2016 |
| B-Raf | Protein kinase | ERK | de la Cova and Greenwald, 2012 |
| Brg1 | Transcription factor | CK1 | Huang et al., 2018 |
| C/EBPα | Transcription factor | – | Bengoechea-Alonso and Ericsson, 2010a |
| C/EBPδ | Transcription factor | GSK3 | Balamurugan et al., 2013 |
| CCDC6 | ATM substrate | – | Zhao et al., 2012 |
| c-JUN | Transcription factor | GSK3 | Nateri et al., 2004; Wei et al., 2005 |
| c-MYB | Transcription factor | GSK3, NLK | Kanei-Ishii et al., 2008; Kitagawa et al., 2009 |
| c-MYC | Transcription factor | GSK3, MAPK | Welcker et al., 2004; Yada et al., 2004 |
| CREB3L1/2 | Transcription factor | – | Yumimoto et al., 2013 |
| Cyclin E | Cyclin protein, Cell cycle | CDK2, GSK3 | Koepp et al., 2001; Moberg et al., 2001; Strohmaier et al., 2001 |
| DEK | Chromatin regulator | GSK3 | Babaei-Jadidi et al., 2011 |
| ERG | TMPRSS2-ERG fusion protein | GSK3, WEE1 | Hong et al., 2020 |
| FAAP20 | Subunit of FA core complex | GSK3 | Wang et al., 2016 |
| Fetuin-A | Alpha-2-HS-glycoprotein | – | Zhao et al., 2018 |
| GCSF-R | Cytokine receptor | GSK3 | Lochab et al., 2013 |
| GRα | Transcription factor | GSK3 | Malyukova et al., 2013 |
| HIF1α | Transcription factor | GSK3 | Cassavaugh et al., 2011 |
| Jun B | Transcription factor | GSK3 | Perez-Benavente et al., 2011 |
| KLF13 | Transcription factor | GSK3 | Kim et al., 2012 |
| KLF2 | Transcription factor | GSK3 | Wang et al., 2013 |
| KLF5 | Transcription factor | GSK3 | Zhao et al., 2010 |
| MCL-1 | Bcl2 family protein | GSK3 | Inuzuka et al., 2011b; Wertz et al., 2011 |
| MED13/13L | Component of mediator complex | – | Davis et al., 2013 |
| mTOR | Protein kinase | – | Mao et al., 2008 |
| NF1 | Ras GTPase regulator | – | Tan et al., 2011 |
| Notch1 | Transcription factor | CDK8 | Fryer et al., 2004 |
| Nrf1 | Transcription factor | GSK3 | Biswas et al., 2011 |
| p100 | Transcription factor | GSK3 | Arabi et al., 2012; Busino et al., 2012; Fukushima et al., 2012 |
| p53 | Transcription factor | GSK3, ATM | Galindo-Moreno et al., 2019; Tripathi et al., 2019; Cui et al., 2020 |
| p63 | Transcription factor | GSK3 | Galli et al., 2010 |
| PDC-1α | Nuclear receptor co-activator | GSK3, p38 | Olson et al., 2008 |
| PLK1 | Serine/threonine kinase | GSK3 | Giraldez et al., 2014 |
| Presenilin | Protease | – | Wu et al., 1998 |
| PU.1 | Transcription factor | GSK3 | Mishra et al., 2021 |
| RCAN1 | Calcineurin A binding protein | – | Lee et al., 2012 |
| REV-ERBα | Nuclear receptor | CDK1 | Zhao et al., 2016 |
| RIG-1 | RNA helicase | – | Wang et al., 2017 |
| SHOC2 | RAS activator | MAPK | Xie et al., 2019 |
| SOX9 | Transcription factor | GSK3 | Hong et al., 2016; Suryo Rahmanto et al., 2016 |
| SRC-3 | Nuclear receptor co-activator | GSK3 | Wu et al., 2007 |
| SREBP1 | Transcription factor | GSK3 | Sundqvist et al., 2005 |
| TG1F1 | Transcription factor | – | Bengoechea-Alonso and Ericsson, 2010b |
| Topo IIα | Topoisomerase | GSK3, CK2 | Chen et al., 2011 |
| TPP1 | Telomere protection protein 1 | GSK3 | Wang et al., 2020 |
| XRCC4 | DNA repair protein | DNA-PKcs | Zhang et al., 2016 |
| ZNF322A | Transcription factor | CK1, GSK3 | Liao et al., 2017 |
| γ-Catenin | Transcription factor | – | Li et al., 2018 |
| ΔNp63α | Transcription factor | GSK3 | Galli et al., 2010 |
In addition to these classical substrates targeted by FBXW7 for ubiquitylation and degradation, several non-canonical substrates were also targeted by FBXW7, but not for degradation (Lan and Sun, 2019). For instance, FBXW7 mediated K63-linked polyubiquitylation of XRCC4 to facilitate the NHEJ repair (Zhang et al., 2016); whereas polyubiquitylation of γ-catenin via K63-linkage by FBXW7 led to enhanced suppression of cell proliferation and G2/M cell cycle transition (Li et al., 2018). Most recently, we found that LSD1 acts as a FBXW7 pseudo-substrate, not being ubiquitylated by FBXW7, but triggering FBXW7 self-ubiquitylation and degradation via disrupting FBXW7 dimerization (Lan et al., 2019). Likewise, EBNA1-binding protein 2 (Ebp2) was also shown to behave as a pseudo-substrate, which directly binds with FBXW7γ in the canonical CPD-dependent manner, not for its degradation, but facilitating the nucleolar localization of FBXW7γ (Welcker et al., 2011).
The Role of FBXW7 in DNA Damage Response
By targeting a wide array of protein substrates for ubiquitylation and degradation, FBXW7 was involved in regulatory networks of various biological processes such as cell cycle progression, cell differentiation, apoptosis, and autophagy (Welcker and Clurman, 2008; Davis et al., 2014). However, knowledge on whether and how FBXW7 regulates DNA damage response (DDR) and repair is rather limited. An early study showed that genetic inactivation of hCDC4 (encoding FBXW7) in karyotype stable colorectal cancer cells resulted in nuclear atypia, such as micronuclei and lobulated or elongated nuclei, as well as chromosomal instability, as evidenced by increased multipolar spindles and euploidy in a manner dependent of cyclin E accumulation (Rajagopalan et al., 2004). However, no rescue experiment nor detailed underlying mechanism was provided to demonstrate whether cyclin E is indeed playing a causal role. Nevertheless, this study did implicate that FBXW7 is likely involved in the maintenance of the genomic integrity. Another in vivo study showed that Fbxw7 is a p53-dependent haploinsufficiency tumor suppressor, and Fbxw7± mice are much more susceptible to radiation-induced tumorigenesis (Mao et al., 2004). A recent study also showed that Fbxw7± mice have an increased the risk of developing gastric cancer induced by chemical carcinogen N-methyl-N-nitrosourea (MNU), which is dependent of the accumulation of DNA damage and c-Myc oncoprotein (Jiang et al., 2017).
Recently, more studies showed that FBXW7 is indeed involved in DDR. Upon DNA damage, FBXW7 was found to promote the ubiquitylation and degradation of several key DDR regulatory proteins, including p53 (Galindo-Moreno et al., 2019; Tripathi et al., 2019; Cui et al., 2020), polo-like kinase 1 (PLK1; Strebhardt, 2010), SOX9 (Hong et al., 2016), and bloom (BLM) helicase (Kharat et al., 2016) among others, which is described below (Figures 3A–D).
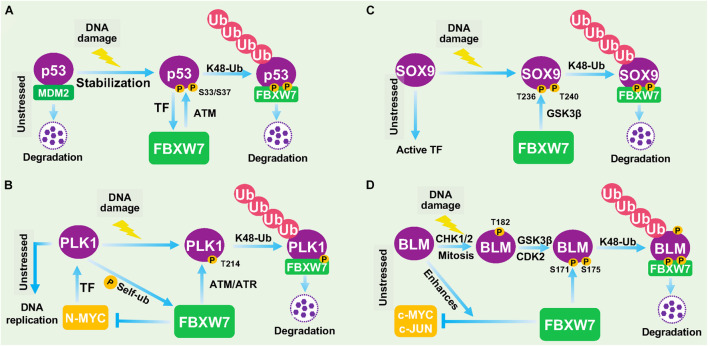
FIGURE 3.
FBXW7 in DNA damage responses. (A) Negative feedback loop between FBXW7 and p53. Under unstressed condition, p53 level is low due to targeted degradation by MDM2 E3 ligase. Upon DNA damage, p53 protein is accumulated to transcriptionally induce FBXW7 expression; the induced FBXW7 then promotes p53 ubiquitylation and degradation after ATM-mediated p53 phosphorylation at Ser33/37 residues (Cui et al., 2020). (B) PLK1 degradation by FBXW7 upon DDR. Under unstressed condition, PLK1 was transcriptionally induced by N-MYC. PLK also phosphorylates FBXW7 to trigger its self-ubiquitylation and cause N-MYC accumulation. Upon DNA damage, ATM/ATR phosphorylates PLK1 at Thr214, which facilitated FBXW7 binding and subsequent ubiquitylation and degradation. (C) FBXW7 degrades SOX9 upon DDR. Under unstressed condition, SOX9 acts as a transcription factor. Upon DNA damage, SOX9 was phosphorylated at Thr236/240 by GSK3β, which facilitated FBXW7 binding and subsequent ubiquitylation and degradation. (D) FBXW7 degrades BLM upon DDR. Upon DNA damage, BLM was sequentially phosphorylated at Thr182 by CHK1/2, and at Thr171 and Ser175 by GSK3β/CDK2, which facilitated FBXW7 binding and subsequent ubiquitylation and degradation. TF, Transcription factor.
Negative Feedback Loop Between FBXW7 and p53
p53 is the best-known tumor suppressor in human cancer, acting as a transcription factor to regulate a wide range of cellular processes, including growth arrest, apoptosis, senescence, DDR, and DNA repair (Vogelstein et al., 2000). The abundance and activity of p53 were fine-tuned by multiple cellular signals and post-translational modifications. In response to DNA damage and other cellular stresses, p53 level is usually increased due to enhanced protein stabilization, mainly resulting from disruption of Mdm2 binding and Mdm2-mediated degradation (Wade et al., 2013). An early study reported that FBXW7 expression was transcriptionally induced upon p53 accumulation after DNA damage stress, thus demonstrating FBXW7 as a bona fide transcriptional target of p53 using cell culture models (Kimura et al., 2003). A subsequent study using a mouse model confirmed that Fbxw7 is indeed a p53-dependent, haploinsufficient tumor suppressor gene (Mao et al., 2004). Very interestingly, three recent studies showed that p53 is also subject to post-translational regulation by FBXW7 for targeted degradation in response to DNA damage in multiple human cancer cell lines (Galindo-Moreno et al., 2019; Tripathi et al., 2019; Cui et al., 2020), thus demonstrating FBXW7 participation in DDR to protect cancer cells from DNA damage-induced cell cycle arrest and apoptosis. Collectively, it is apparent that a negative feedback loop exists between FBXW7 and p53. In response to DNA damage, p53 protein is accumulated to transcriptionally induce FBXW7 expression; the induced FBXW7 then promotes p53 degradation to keep the p53 level in check as a mechanism of self-defense (Figure 3A). Given both FBXW7 and p53 are frequently mutated in many types of human cancers, we performed a bioinformatics analysis on TCGA databases and found interestingly that mutations of p53 and FBXW7 in human cancers are co-occurrence (Zehir et al., 2017), suggesting that this p53-FBXW7 negative feedback loop may have a biological implication.
FBXW7 Degrades PLK1 in Response to DNA Damage
PLK1 is a serine/threonine-protein kinase that performs important biological functions mainly in the late G2/M phase of cell cycle, including centrosome maturation, spindle assembly and sister chromatid separation (Strebhardt, 2010). PLK1 was also shown to promote DNA replication by regulating pre-replicative complexes (pre-RCs) loading of mini-chromosome maintenance (MCM) 2/6 (Yoo et al., 2004; Tsvetkov and Stern, 2005). In response to UV-induced DNA damage, PLK1 was degraded by FBXW7 in cells arrested at G1- and S-phase, thus blocking the formation of pre-RCs to prevent the improper progression of cell cycle and avoid the proliferation of cells carrying damaged DNA (Giraldez et al., 2014). Thus, by degrading PLK1 to temporarily halt cell cycle progression, FBXW7 acts as a gate-keeper to ensure genome stability (Giraldez et al., 2014; Figure 3B).
FBXW7 Degrades SOX9 in Response to DNA Damage
SOX9 is a member of the high-mobility group (HMG)-box class of transcription factors, and plays a key role in chondrocytes differentiation and skeletal development (Adam et al., 2015). It was reported that under the DNA damage induced by UV irradiation or genotoxic chemotherapeutic agents, SOX9 was actively degraded in various cancer types and even in normal epithelial cells. Mechanistic study revealed that FBXW7 is the E3 ubiquitin ligase mediating SOX9 degradation in a manner dependent on prior phosphorylation by GSK3β (Hong et al., 2016; Figure 3C). However, how FBXW7-mediated SOX9 degradation contributes to overall DDR remains elusive. Furthermore, SOX9 protein was also targeted by FBXW7 for proteasomal degradation in medulloblastoma cells even under normal unstressed conditions (Suryo Rahmanto et al., 2016), suggesting that FBXW7-mediated SOX9 degradation might be in a cell and context dependent event, and not specific to DDR.
FBXW7 Degrades Bloom Helicase in Response to DNA Damage
BLM is an ATP-dependent DNA helicase that unwinds single- and double-stranded DNA. Once stalled DNA replication or DNA damage occurs, BLM is recruited to participate in fixing the genomic error (Chu and Hickson, 2009). The interaction between BLM and FBXW7 has been previously reported to enhance FBXW7-mediated c-MYC degradation (Chandra et al., 2013). A more recent study showed that the protein levels of BLM is dynamically fluctuant during cell cycle progression with the interphase cells having a higher level than the mitotic cells. Further mechanistic studies revealed that FBXW7 promoted the K48-linked polyubiquitylation of BLM in a manner dependent on GSK3β and CDK2 -mediated prior phosphorylation of BLM at Thr171 and Ser175 (Kharat et al., 2016). The authors further found that FBXW7-promoted BLM degradation is a mitosis-specific event requiring prior phosphorylation at Thr182 by CHK1/CHK2 (Figure 3D). Given that CHK1/CHK2 activation is a common signal during DDR, FBXW7-mediated BLM degradation triggered by CHK1/CHK2 is likely involved in the process of DDR, although detailed underlying mechanism remains elusive.
Furthermore, we recently found that FBXW7 was recruited to the DSB sites by poly(ADP-ribose) (PAR) (Zhang Q. et al., 2019) and maintained at DNA damage sites in a ATM-dependent manner upon laser irradiation to facilitate the non-homolog end-joining (NHEJ) repair (Zhang et al., 2016) (see below). The others reported that FBXW7 binds to telomere protection protein 1 (TPP1) and promotes its polyubiquitylation at multi-sites for enhanced degradation, which triggers telomere uncapping and DNA damage response and affects senescence and fibrosis of pulmonary epithelial stem cell (Wang et al., 2020). Upon DNA damage stress in prostate cancer (PCa) cells, TMPRSS2-ERG gene fusion product was degraded by FBXW7 in a manner dependent of GSK3β and WEE1 kinases. Blockage of such degradation promoted genotoxic therapy-resistant growth of fusion-positive PCa cells both in vitro and in vivo (Hong et al., 2020). In nasopharyngeal carcinoma cells, fructose-1,6-bisphosphatase 1 (FBP1) was found to suppress the autoubiquitylation of FBXW7 to restrained mTOR-glycolysis signals and promote radiation-induced apoptosis and DNA damage (Zhang et al., 2021). FBXW7 also cooperated with MDM2 following DDR, to regulate the levels of the pro-proliferative ΔNp63α protein, resulting in cell proliferation (Galli et al., 2010). Finally, in addition to p53, other transcription factors known to be FBXW7 substrates were also reported to be involved in DDR. For example, HIF-1 was involved in γH2AX accumulation by tumor hypoxia (Wrann et al., 2013), whereas KLF5 plays a significant role in the DNA damage response by regulating the phosphorylation of CHK1/2 (Zhang H. et al., 2019). Taken together, it appears that FBXW7 is indeed implicated in DDR that likely contribute to its role in the maintenance of genome integrity, mainly by targeted ubiquitylation and degradation of key regulatory proteins, such as p53, PLK1, and BLM.
The Role of FBXW7 in DNA Damage Repair
The DDR and repairs are two sequential events which are essential for the maintenance of genomic stability. The DNA attack by different external and internal insults produces a variety of DNA lesion modalities, mainly including simple base modification, base mismatches, bulky DNA adducts, inter-strand and intra-strand crosslinks (ICLs), SSBs and DSBs, which trigger different types of DDRs and repair processes to fix these damages (Roos et al., 2016). For example, base mismatches are repaired by mismatch repair machinery (MMR), ICLs are repaired by NER, HR, and Fanconi Anaemia (FA) repair pathways, whereas DSBs are mainly repaired by NHEJ or HR (Roos et al., 2016). So far, FBXW7 was found to regulate FA and NHEJ pathways by targeting Fanconi anemia core complex-associated protein (FAAP) 20 (Wang et al., 2016, 2019) and XRCC4 (Zhang et al., 2016), respectively (Figure 4).
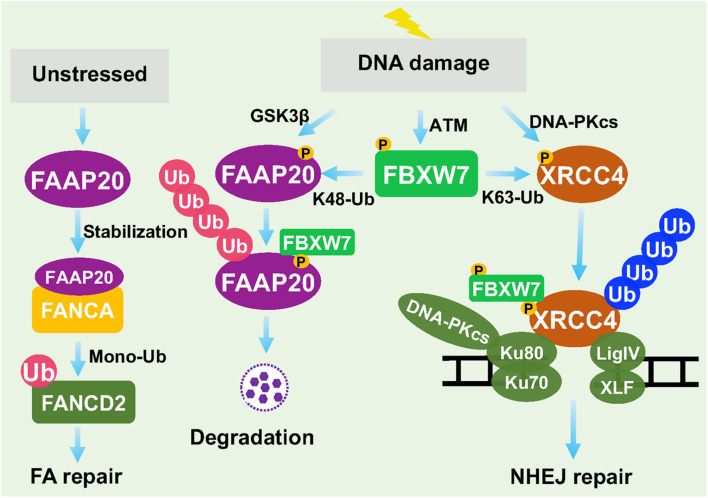
FIGURE 4.
FBXW7 in regulation of DNA repair. Under unstressed condition, FAAP20 binds FANCA to facilitate mono-ubiquitylation of FANCD2. Upon DDR, FAAP20 was phosphorylated by GSK3β to facilitate FBXW7 binding and ubiquitylation. On the other hand, DNA-PKcs phosphorylated XRCC4, whereas ATM phosphorylated FBXW7. At the damage site, FBXW7 promoted XRCC4 polyubiquitylation via the K63 linkage to facilitate recruitment of Ku70/Ku80 for NHEJ repair.
In Regulation of Fanconi Anemia Pathway
The Fanconi Anemia (FA) pathway is a DNA repair process responsible for resolving ICLs (Roos et al., 2016). Germ-line mutations of key FA genes caused inherited FA disorders with cancer predisposition. The key step of FA pathway to initiate the repair process is the monoubiquitylation of FANCD2, mediated by FA core complex containing a scaffold protein FANCA. FANCA has a binding partner, FAAP20, which is required for its stability (Leung et al., 2012). It was reported that FBXW7 promoted the polyubiquitylation of FAAP20 in a manner dependent on GSK3β-mediated prior phosphorylation at Ser113 in response to DNA damage. Thus, by modulating the levels of FAAP20 and subsequent stability of FANCA, FBXW7 acts as a upstream regulator of FA pathway and its repair process (Wang et al., 2016).
In Regulation of Non-homologous End Joining Repair
Recently, we reported that in response to DSB DNA damage, ATM is activated to remain FBXW7 at the damage sites and then trigger NHEJ, but not HR, repair (Zhang et al., 2016). Specifically, ionizing radiation caused DNA damage that activates both DNA-PKcs and ATM. On the one hand, DNA-PKcs phosphorylates XRCC4, a key regulator of NHEJ on damaged site, and on the other hand, PAR and ATM recruit FBXW7 to the DSBs sites, where FBXW7 promotes polyubiquitylation of phosphorylated XRCC4 via the K63 linkage. Polyubiquitylated XRCC4 was not delivered to proteasome for degradation, rather to build up a platform that facilitated the recruitment of Ku70/80 heterodimer to promote NHEJ repair (Zhang et al., 2016; Zhang Q. et al., 2019). Thus, FBXW7 is actively involved in processes of DNA damage repair by FA and NHEJ.
Furthermore, it is worth noting that c-MYC, as a typical substrate of FBXW7, was also reported to function in DNA damage repair. Specifically, it was reported that c-MYC suppresses DSB accumulation in a manner strictly dependent of Polymerase Associated Factor 1 complex (Endres et al., 2021). Another study reported that c-MYC directly interacts with Ku70 protein through its Myc box II (MBII) domain to block DSB repair and V(D)J recombination, which probably occur through inhibition of the NHEJ pathway (Li et al., 2012). Given the fact that FBXW7 has a variety of substrates, it is likely that some of these substrates may regulate DNA damage repair in an indirect manner, if not in a direct manner, which is, however, out of scope of this focused review.
Conclusion and Future Perspectives
In the past two decades, the FBXW7-related studies were mainly focused on identification and characterization of its substrates. To date, more than 40 proteins have been identified, and most of them are transcription factors that regulate a broad range of biological processes. By timely modulating ubiquitylation and degradation of these substrates, FBXW7 acts as a central regulator of key biological processes and various cellular signal pathways. However, the knowledge on the role of FBXW7 in DDR and repair is rather limited, though the involvement of FBXW7 in genomic stability was implicated almost two decades ago. In this review, we summarized current available data and concluded that FBXW7 is indeed actively involved in DDR and repair processes by acting as an E3 ligase to promote ubiquitylation of (a) key DDR regulatory proteins for degradation or (b) key repair proteins for facilitating repair process (Figures 3, 4).
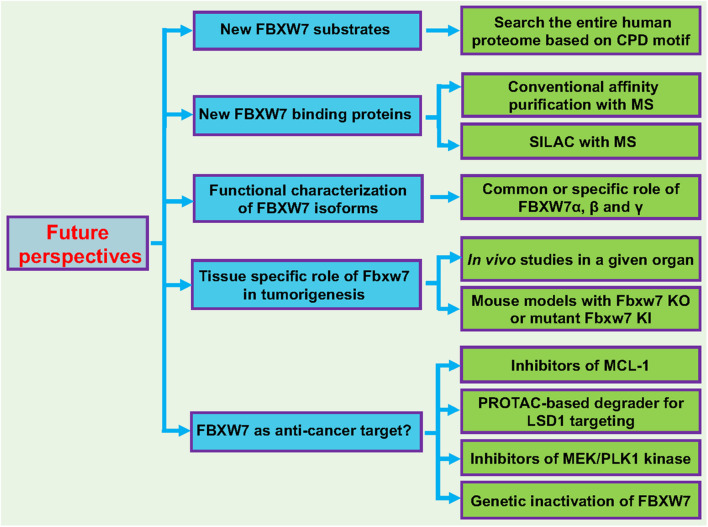
Here, we propose few future studies on FBXW7 to further elucidate the role of FBXW7 in a variety of biological processes (Figure 5).
FIGURE 5.
Future perspectives. Five future directions are proposed to further broaden our understanding of FBXW7 functions: (1) Identification and characterization of new FBXW7 substrates; (2) Identification and characterization of FBXW7 binding proteins; (3) Functional characterization of FBXW7 isoforms (α, β, and γ); (4) Tissue specific role of Fbxw7 in tumorigenesis using mouse cKO or KI models; (5) Target human cancers by reactivating FBXW7. See text for details.
Identification and Characterization of Additional FBXW7 Substrates Which May or May Not Be Involved in DNA Damage Response and Repair Processes
Given that FBXW7 binding motif, also known as CDC4 phospho-degron (CPD), on a substrate has been well-defined, the quick and easy way to identify new putative FBXW7 substrate candidates is to search the entire human proteome for CPD motif or its modified version with mimicking negative charged residues within the CPD motif, followed by detailed characterization of candidates as the bona fide FBXW7 substrates. FBXW7 functions will be further extended, based upon the known functions of these newly identified substrates in a given signaling pathway.
Identification and Characterization of FBXW7 Binding Proteins That Regulate FBXW7 Functions
Conventional affinity purification coupled with Mass-Spectrometry (MS) or stable isotope labeling by amino acids in cell culture (SILAC) with the MS-based quantitative proteomics (Ong and Mann, 2006) can be used to identify FBXW7 binding proteins under unstressed physiological condition or after a given stress of interest. The identified binding proteins can also be FBXW7 substrates or merely FBXW7 binding proteins not subjected to FBXW-mediated ubiquitylation. FBXW7 may have novel functions independent of its E3 ligase activity, which can be identified and defined through the characterization of these binding partners.
Functional Characterization of FBXW7 Isoforms
The majority of current studies have been focused upon FBXW7α localized in the nucleus. No detailed functional characterization of other isoforms, particularly FBXW7γ localized in the nucleolus. It is unclear whether other SCF components are localized in nucleolus and if not, FBXW7γ may not act as an active E3 ligase. It will be interesting to define the role of this isoform, particularly in response to ribosomal stress that is mainly triggered in nucleolus (Golomb et al., 2014), a subcellular organelle which also plays a role in maintenance of genome stability (Lindstrom et al., 2018).
Tissue Specific Role of Fbxw7 in Tumorigenesis
In human cancer cells, FBXW7 is frequently inactivated via point mutations, allele deletion, promoter methylation, and induced self-ubiquitylation (Welcker and Clurman, 2008; Wang et al., 2012; Lan and Sun, 2019). It is very likely that cancer cells with FBXW7 inactivation have growth advantage and were selected during tumorigenesis. However, these correlation-based studies did not validate whether FBXW7 truly plays a causal role in organ-specific tumorigenesis. Thus, for in vivo validation of the role of FBXW7 in tumorigenesis in a given organ, the mouse models with tissue-specific Fbxw7 KO or mutant Fbxw7 KI should be generated, particularly in those tissues with high frequency of FBXW7 alterations (Figure 2A). A detailed characterization of these genetically modified mouse models will reveal whether FBXW7 indeed plays a driver role or merely co-operate with other dominant oncogenes (such as Kras activation) or tumor suppressors (such as loss of p53 or Pten) in compound mouse models in a particular organ.
FBXW7 as a Potential Anti-cancer Target?
FBXW7 is a tumor suppressor and FBXW7 itself certainly cannot be served as a direct anti-cancer target. However, few correlation studies have shown an association of FBXW7 expression with chemotherapeutic sensitivity. For example, loss of FBXW7 was associated with increased sensitivity of lung cancer cells to histone deacetylase (HDAC) inhibitor, MS-275 (Yokobori et al., 2014), and tumor cell lines harboring deletions or mutations in FBXW7 are particularly sensitive to rapamycin treatment (Mao et al., 2008). Given the fact that pro-survival protein MCL-1 is a substrate of FBXW7, it is not surprised that T-cell acute lymphoblastic leukemia cell lines with defective FBXW7 have increased levels of MCL1 and are particularly resistance to BCL2 antagonist ABT-737 in a manner dependent of MCL1 (Inuzuka et al., 2011a). Furthermore, FBXW7 loss conferred resistance to anti-tubulin agents and promoted chemotherapeutic-induced polyploidy due to MCL1 accumulation (Wertz et al., 2011). These studies together suggest that loss-of-function FBXW7 mutations indeed impact chemotherapy sensitivity, although it is mainly due to accumulation of its anti-apoptotic substrates, not directly related to its DNA repair function. Nevertheless, small molecule inhibitors of MCL1, currently under clinical development (Hird and Tron, 2019; Negi and Murphy, 2021), are certainly a proper choice to target human cancers with FBXW7 mutations.
On the other hand, the upstream regulators of FBXW7 also provide sound strategies for potential FBXW7-based translational application in cancer treatment. Specifically, several oncoproteins have been shown to trigger FBXW7 self-ubiquitylation and targeting these oncoproteins would, in theory, stabilize FBXW7 to execute its tumor suppressor function.
The first case is LSD1 (lysine-specific demethylase-1), an enzyme overexpressed in many human cancers with correlation of poor patient survival (Kooistra and Helin, 2012), which has been validated as an attractive cancer target with extensive drug discovery efforts (Fu et al., 2017). We recently found that LSD1 is a pseudo FBXW7 substrate, which binds with FBXW7 in the classical CPD-dependent manner. Instead of being ubiquitylated by FBXW7 for proteasomal degradation, LSD1-FBXW7 binding inhibited FBXW7 dimerization, leading to FBXW7 self-ubiquitylation and subsequent degradation via proteasome and lysosome systems in a manner independent of its demethylase activity (Lan et al., 2019; Gu et al., 2020). Currently, several LSD1 demethylase inhibitors have been in the Phase I/IIa clinical trials (Hosseini and Minucci, 2017). Future drug discovery effort could be directed to screen for small molecules that specifically bind to LSD1, not necessary to inhibit its demethylase activity, followed by discovery of PROTAC-based degrader (Gu et al., 2018) for LSD1 targeting. This type of LSD1 specific PROTAC drugs should have broad applications for the treatment of human cancers harboring a wild type FBXW7 with LSD1 overexpression by reactivating FBXW7, as well as for potential immunotherapy in combination with PD-L1 blockade (Sheng et al., 2018).
The second example is the ERK kinase that has been shown to phosphorylate FBXW7, leading to its destabilization in a manner dependent of PIN-1 (Ji et al., 2015). The inhibitors of MEK, an ERK upstream kinase, have also been in a number of Phase II/III clinical trials (Kim and Giaccone, 2018). The third case is the PLK kinase that has also been shown to phosphorylate FBXW7 to promote its self-ubiquitylation (Xiao et al., 2016). Again, several PLK inhibitors were also under clinic development (Janning and Fiedler, 2014). These inhibitors may have new applications in the treatment of human cancers with activated MAPK or PLK1 signals by reactivating FBXW7. Finally, in a subset of human cancers with developed resistance to chemo- and radiotherapies, but harboring wild-type p53, inactivation of FBXW7 via genetic approaches may reactivate p53 to overcome the resistance (Cui et al., 2020).
Author Contributions
HL wrote the first draft of the manuscript. YS revised and finalized the manuscript. Both authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Haomin Li at the Children’s Hospital of Zhejiang University School of Medicine for his bioinformatics analysis on association of mutations of p53 and FBXW7 in human cancers.
Footnotes
Funding
This work was supported by the National Key R&D Program of China (2016YFA0501800 to YS), National Natural Science Foundation of China (82102947 to HL), and National Natural Science Foundation of China (81630076 to YS).
References
- Adam R. C., Yang H., Rockowitz S., Larsen S. B., Nikolova M., Oristian D. S., et al. (2015). Pioneer factors govern super-enhancer dynamics in stem cell plasticity and lineage choice. Nature 521:366. 10.1038/nature14289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabi A., Ullah K., Branca R. M. M., Johansson J., Bandarra D., Haneklaus M., et al. (2012). Proteomic screen reveals Fbw7 as a modulator of the NF-kappa B pathway. Nat. Commun. 3:976. 10.1038/ncomms1975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babaei-Jadidi R., Li N. N., Saadeddin A., Spencer-Dene B., Jandke A., Muhammad B., et al. (2011). FBXW7 influences murine intestinal homeostasis and cancer, targeting Notch, Jun, and DEK for degradation. J. Exp Med. 208 295–312. 10.1084/jem.20100830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahram F., von der Lehr N., Cetinkaya C., Larsson L. G. (2000). c-Myc hot spot mutations in lymphomas result in inefficient ubiquitination and decreased proteasome-mediated turnover. Blood 95 2104–2110. [PubMed] [Google Scholar]
- Balamurugan K., Sharan S., Klarmann K. D., Zhang Y., Coppola V., Summers G. H., et al. (2013). FBXW7 alpha attenuates inflammatory signalling by downregulating C/EBP delta and its target gene Tlr4. Nat. Commun. 4:1662. 10.1038/ncomms2677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea-Alonso M. T., Ericsson J. (2010a). The ubiquitin ligase Fbxw7 controls adipocyte differentiation by targeting C/EBP alpha for degradation. Proc. Natl. Acad. Sci. U.S.A. 107 11817–11822. 10.1073/pnas.0913367107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bengoechea-Alonso M. T., Ericsson J. (2010b). Tumor suppressor Fbxw7 regulates TGF beta signaling by targeting TGIF1 for degradation. Oncogene 29 5322–5328. 10.1038/onc.2010.278 [DOI] [PubMed] [Google Scholar]
- Biswas M., Phan D., Watanabe M., Chan J. Y. (2011). The Fbw7 tumor suppressor regulates nuclear factor E2-related factor 1 transcription factor turnover through proteasome-mediated proteolysis. J. Biol. Chem. 286 39282–39289. 10.1074/jbc.M111.253807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busino L., Millman S. E., Scotto L., Kyratsous C. A., Basrur V., O’Connor O., et al. (2012). Fbxw7 alpha- and GSK3-mediated degradation of p100 is a pro-survival mechanism in multiple myeloma. Nat. Cell Biol. 14:375. 10.1038/ncb2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassavaugh J. M., Hale S. A., Wellman T. L., Howe A. K., Wong C., Lounsbury K. M. (2011). Negative regulation of HIF-1 alpha by an FBW7-mediated degradation pathway during hypoxia. J. Cell Biochem. 112 3882–3890. 10.1002/jcb.23321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra S., Priyadarshini R., Madhavan V., Tikoo S., Hussain M., Mudgal R., et al. (2013). Enhancement of c-Myc degradation by BLM helicase leads to delayed tumor initiation. J. Cell Sci. 126(Pt 16) 3782–3795. 10.1242/jcs.124719 [DOI] [PubMed] [Google Scholar]
- Chen M. C., Chen C. H., Chuang H. C., Kulp S. K., Teng C. M., Chen C. S. (2011). Novel mechanism by which histone deacetylase inhibitors facilitate topoisomerase II alpha degradation in hepatocellular carcinoma cells. Hepatology 53 148–159. 10.1002/hep.23964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu W. K., Hickson I. D. (2009). RecQ helicases: multifunctional genome caretakers. Nat. Rev. Cancer. 9 644–654. [DOI] [PubMed] [Google Scholar]
- Ciechanover A. (1998). The ubiquitin-proteasome pathway: on protein death and cell life. EMBO J. 17 7151–7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D., Xiong X., Shu J., Dai X., Sun Y., Zhao Y. (2020). FBXW7 confers radiation survival by targeting p53 for degradation. Cell Rep. 30 497–509.e4. 10.1016/j.celrep.2019.12.032 [DOI] [PubMed] [Google Scholar]
- Davis M. A., Larimore E. A., Fissel B. M., Swanger J., Taatjes D. J., Clurman B. E. (2013). The SCF-Fbw7 ubiquitin ligase degrades MED13 and MED13L and regulates CDK8 module association with Mediator. Gene Dev. 27 151–156. 10.1101/gad.207720.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis R. J., Welcker M., Clurman B. E. (2014). Tumor suppression by the Fbw7 ubiquitin ligase: mechanisms and opportunities. Cancer Cell 26 455–464. 10.1016/j.ccell.2014.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cova C., Greenwald I. (2012). SEL-10/Fbw7-dependent negative feedback regulation of LIN-45/Braf signaling in C. elegans via a conserved phosphodegron. Gene Dev. 26 2524–2535. 10.1101/gad.203703.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durgan J., Parker P. J. (2010). Regulation of the tumour suppressor Fbw7 alpha by PKC-dependent phosphorylation and cancer-associated mutations. Biochem. J. 432 77–87. 10.1042/BJ20100799 [DOI] [PubMed] [Google Scholar]
- Endres T., Solvie D., Heidelberger J. B., Andrioletti V., Baluapuri A., Ade C. P., et al. (2021). Ubiquitylation of MYC couples transcription elongation with double-strand break repair at active promoters. Mol. Cell 81 830–844.e13. 10.1016/j.molcel.2020.12.035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn K., Lowndes N. F., Grenon M. (2012). Eukaryotic DNA damage checkpoint activation in response to double-strand breaks. Cell Mol. Life Sci. 69 1447–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryer C. J., White J. B., Jones K. A. (2004). Mastermind recruits CycC : CDK8 to phosphorylate the notch ICD and coordinate activation with turnover. Mol. Cell 16 509–520. 10.1016/j.molcel.2004.10.014 [DOI] [PubMed] [Google Scholar]
- Fu X., Zhang P., Yu B. (2017). Advances toward LSD1 inhibitors for cancer therapy. Future Med. Chem. 9 1227–1242. [DOI] [PubMed] [Google Scholar]
- Fukushima H., Matsumoto A., Inuzuka H., Zhai B., Lau A. W., Wan L. X., et al. (2012). SCFFbw7 modulates the NF kappa B signaling pathway by targeting NF kappa B2 for ubiquitination and destruction. Cell Rep. 1 434–443. 10.1016/j.celrep.2012.04.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo-Moreno M., Giraldez S., Limon-Mortes M. C., Belmonte-Fernandez A., Reed S. I., Saez C., et al. (2019). SCF(FBXW7)-mediated degradation of p53 promotes cell recovery after UV-induced DNA damage. FASEB J. 33 11420–11430. 10.1096/fj.201900885R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli F., Rossi M., D’Alessandra Y., De Simone M., Lopardo T., Haupt Y., et al. (2010). MDM2 and Fbw7 cooperate to induce p63 protein degradation following DNA damage and cell differentiation. J. Cell. Sci. 123 2423–2433. 10.1242/jcs.061010 [DOI] [PubMed] [Google Scholar]
- Giraldez S., Herrero-Ruiz J., Mora-Santos M., Japon M. A., Tortolero M., Romero F. (2014). SCF(FBXW7alpha) modulates the intra-S-phase DNA-damage checkpoint by regulating Polo like kinase-1 stability. Oncotarget 5 4370–4383. 10.18632/oncotarget.2021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golomb L., Volarevic S., Oren M. (2014). p53 and ribosome biogenesis stress: the essentials. FEBS Lett. 588 2571–2579. [DOI] [PubMed] [Google Scholar]
- Gu F., Lin Y., Wang Z., Wu X., Ye Z., Wang Y., et al. (2020). Biological roles of LSD1 beyond its demethylase activity. Cell Mol. Life Sci. 77 3341–3350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu S., Cui D., Chen X., Xiong X., Zhao Y. (2018). PROTACs: an emerging targeting technique for protein degradation in drug discovery. Bioessays 40:e1700247. [DOI] [PubMed] [Google Scholar]
- Hao B., Oehlmann S., Sowa M. E., Harper J. W., Pavletich N. P. (2007). Structure of a Fbw7-Skp1-cyclin E complex: multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol Cell. 26 131–143. 10.1016/j.molcel.2007.02.022 [DOI] [PubMed] [Google Scholar]
- Hershko A., Ciechanover A., Varshavsky A. (2000). Basic medical research award. the ubiquitin system. Nat. Med. 6 1073–1081. [DOI] [PubMed] [Google Scholar]
- Hird A. W., Tron A. E. (2019). Recent advances in the development of Mcl-1 inhibitors for cancer therapy. Pharmacol. Ther. 198 59–67. [DOI] [PubMed] [Google Scholar]
- Hong X., Liu W., Song R., Shah J. J., Feng X., Tsang C. K., et al. (2016). SOX9 is targeted for proteasomal degradation by the E3 ligase FBW7 in response to DNA damage. Nucleic Acids Res. 44 8855–8869. 10.1093/nar/gkw748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong Z., Zhang W., Ding D. L., Huang Z. L., Yan Y. Q., Cao W., et al. (2020). DNA damage promotes TMPRSS2-ERG oncoprotein destruction and prostate cancer suppression via signaling converged by GSK3 beta and WEE1. Mol. Cell 79:1008. 10.1016/j.molcel.2020.07.028 [DOI] [PubMed] [Google Scholar]
- Hosseini A., Minucci S. (2017). A comprehensive review of lysine-specific demethylase 1 and its roles in cancer. Epigenomics 9 1123–1142. [DOI] [PubMed] [Google Scholar]
- Huang L. Y., Zhao J. J., Chen H., Wan L. X., Inuzuka H., Guo J. P., et al. (2018). SCFFBW7-mediated degradation of Brg1 suppresses gastric cancer metastasis. Nat. Commun. 9:3569. 10.1038/s41467-018-06038-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H., Gao D., Finley L. W., Yang W., Wan L., Fukushima H., et al. (2012). Acetylation-dependent regulation of Skp2 function. Cell 150 179–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H., Shaik S., Onoyama I., Gao D., Tseng A., Maser R. S., et al. (2011a). SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471 104–109. 10.1038/nature09732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inuzuka H., Shaik S., Onoyama I., Gao D. M., Tseng A., Maser R. S., et al. (2011b). SCFFBW7 regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 471 104–U28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janning M., Fiedler W. (2014). Volasertib for the treatment of acute myeloid leukemia: a review of preclinical and clinical development. Future Oncol. 10 1157–1165. [DOI] [PubMed] [Google Scholar]
- Ji S., Qin Y., Shi S., Liu X., Hu H., Zhou H., et al. (2015). ERK kinase phosphorylates and destabilizes the tumor suppressor FBW7 in pancreatic cancer. Cell Res. 25 561–573. 10.1038/cr.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y. N., Qi X. M., Liu X. Y., Zhang J., Ji J., Zhu Z. G., et al. (2017). Fbxw7 haploinsufficiency loses its protection against DNA damage and accelerates MNU-induced gastric carcinogenesis. Oncotarget 8 33444–33456. 10.18632/oncotarget.16800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J., Cardozo T., Lovering R. C., Elledge S. J., Pagano M., Harper J. W. (2004). Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 18 2573–2580. 10.1101/gad.1255304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanei-Ishii C., Nomura T., Takagi T., Watanabe N., Nakayama K. I., Ishii S. (2008). Fbxw7 Acts as an E3 ubiquitin ligase that targets c-Myb for Nemo-like Kinase (NLK)-induced degradation. J. Biol. Chem. 283 30540–30548. 10.1074/jbc.M804340200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kharat S. S., Tripathi V., Damodaran A. P., Priyadarshini R., Chandra S., Tikoo S., et al. (2016). Mitotic phosphorylation of Bloom helicase at Thr182 is required for its proteasomal degradation and maintenance of chromosomal stability. Oncogene 35 1025–1038. 10.1038/onc.2015.157 [DOI] [PubMed] [Google Scholar]
- Kim C., Giaccone G. (2018). MEK inhibitors under development for treatment of non-small-cell lung cancer. Expert Opin. Investig. Drugs 27 17–30. [DOI] [PubMed] [Google Scholar]
- Kim D. S., Zhang W., Millman S. E., Hwang B. J., Kwon S. J., Clayberger C., et al. (2012). Fbw7 gamma-mediated degradation of KLF13 prevents RANTES expression in resting human but not murine T lymphocytes. Blood 120 1658–1667. 10.1182/blood-2012-03-415968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T., Gotoh M., Nakamura Y., Arakawa H. (2003). hCDC4b, a regulator of cyclin E, as a direct transcriptional target of p53. Cancer Sci. 94 431–436. 10.1111/j.1349-7006.2003.tb01460.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitagawa K., Hiramatsu Y., Uchida C., Isobe T., Hattori T., Oda T., et al. (2009). Fbw7 promotes ubiquitin-dependent degradation of c-Myb: involvement of GSK3-mediated phosphorylation of Thr-572 in mouse c-Myb. Oncogene 28 2393–2405. 10.1038/onc.2009.111 [DOI] [PubMed] [Google Scholar]
- Koepp D. M., Schaefer L. K., Ye X., Keyomarsi K., Chu C., Harper J. W., et al. (2001). Phosphorylation-dependent ubiquitination of cyclin E by the SCFFbw7 ubiquitin ligase. Science 294 173–177. [DOI] [PubMed] [Google Scholar]
- Kooistra S. M., Helin K. (2012). Molecular mechanisms and potential functions of histone demethylases. Nat. Rev. Mol. Cell Biol. 13 297–311. [DOI] [PubMed] [Google Scholar]
- Kwon Y. W., Kim I. J., Wu D., Lu J., Stock W. A., Liu Y. Y., et al. (2012). Pten regulates aurora-A and cooperates with Fbxw7 in modulating radiation-induced tumor development. Mol. Cancer Res. 10 834–844. 10.1158/1541-7786.MCR-12-0025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H., Tan M., Zhang Q., Yang F., Wang S., Li H., et al. (2019). LSD1 destabilizes FBXW7 and abrogates FBXW7 functions independent of its demethylase activity. Proc. Natl. Acad. Sci. U.S.A. 116 12311–12320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan H. Y., Sun Y. (2019). FBXW7 E3 ubiquitin ligase: degrading, not degrading, or being degraded. Protein Cell 10 861–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J. W., Kang H. S., Lee J. Y., Lee E. J., Rhim H., Yoon J. H., et al. (2012). The transcription factor STAT2 enhances proteasomal degradation of RCAN1 through the ubiquitin E3 ligase FBW7. Biochem. Biophys. Res. Commun. 420 404–410. 10.1016/j.bbrc.2012.03.007 [DOI] [PubMed] [Google Scholar]
- Leung J. W., Wang Y., Fong K. W., Huen M. S., Li L., Chen J. (2012). Fanconi anemia (FA) binding protein FAAP20 stabilizes FA complementation group A (FANCA) and participates in interstrand cross-link repair. Proc. Natl. Acad. Sci. U.S.A. 109 4491–4496. 10.1073/pnas.1118720109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Hu K., Xiao X., Wu W., Yan H., Chen H., et al. (2018). FBW7 suppresses cell proliferation and G2/M cell cycle transition via promoting gamma-catenin K63-linked ubiquitylation. Biochem. Biophys. Res. Commun. 497 473–479. 10.1016/j.bbrc.2018.01.192 [DOI] [PubMed] [Google Scholar]
- Li Z., Owonikoko T. K., Sun S. Y., Ramalingam S. S., Doetsch P. W., Xiao Z. Q., et al. (2012). c-Myc suppression of DNA double-strand break repair. Neoplasia 14 1190–1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao S. Y., Chiang C. W., Hsu C. H., Chen Y. T., Jen J., Juan H. F., et al. (2017). CK1 delta/GSK3 beta/FBXW7 alpha axis promotes degradation of the ZNF322A oncoprotein to suppress lung cancer progression. Oncogene 36 5722–5733. 10.1038/onc.2017.168 [DOI] [PubMed] [Google Scholar]
- Lindstrom M. S., Jurada D., Bursac S., Orsolic I., Bartek J., Volarevic S. (2018). Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 37 2351–2366. 10.1038/s41388-017-0121-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lochab S., Pal P., Kapoor I., Kanaujiya J. K., Sanyal S., Behre G., et al. (2013). E3 ubiquitin ligase Fbw7 negatively regulates granulocytic differentiation by targeting G-CSFR for degradation. Biochim. Biophys. Acta. 1833 2639–2652. 10.1016/j.bbamcr.2013.06.018 [DOI] [PubMed] [Google Scholar]
- Malyukova A., Brown S., Papa R., O’Brien R., Giles J., Trahair T. N., et al. (2013). FBXW7 regulates glucocorticoid response in T-cell acute lymphoblastic leukaemia by targeting the glucocorticoid receptor for degradation. Leukemia 27 1053–1062. 10.1038/leu.2012.361 [DOI] [PubMed] [Google Scholar]
- Mao J. H., Kim I. J., Wu D., Climent J., Kang H. C., DelRosario R., et al. (2008). FBXW7 targets mTOR for degradation and cooperates with PTEN in tumor suppression. Science 321 1499–1502. 10.1126/science.1162981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao J. H., Perez-Losada J., Wu D., Delrosario R., Tsunematsu R., Nakayama K. I., et al. (2004). Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 432 775–779. 10.1038/nature03155 [DOI] [PubMed] [Google Scholar]
- Matsumoto A., Onoyama I., Nakayama K. I. (2006). Expression of mouse Fbxw7 isoforms is regulated in a cell cycle- or p53-dependent manner. Biochem. Biophys. Res. Commun. 350 114–119. [DOI] [PubMed] [Google Scholar]
- Min S. H., Lau A. W., Lee T. H., Inuzuka H., Wei S., Huang P., et al. (2012). Negative regulation of the stability and tumor suppressor function of Fbw7 by the Pin1 prolyl isomerase. Mol. Cell. 46 771–783. 10.1016/j.molcel.2012.04.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra M., Thacker G., Sharma A., Singh A. K., Upadhyay V., Sanyal S., et al. (2021). FBW7 inhibits myeloid differentiation in acute myeloid leukemia via GSK3-dependent ubiquitination of PU.1. Mol. Cancer Res. 19 261–273. 10.1158/1541-7786.MCR-20-0268 [DOI] [PubMed] [Google Scholar]
- Mo J. S., Ann E. J., Yoon J. H., Jung J., Choi Y. H., Kim H. Y., et al. (2011). Serum- and glucocorticoid-inducible kinase 1 (SGK1) controls Notch1 signaling by downregulation of protein stability through Fbw7 ubiquitin ligase. J. Cell Sci. 124(Pt 1) 100–112. 10.1242/jcs.073924 [DOI] [PubMed] [Google Scholar]
- Moberg K. H., Bell D. W., Wahrer D. C. R., Haber D. A., Hariharan I. K. (2001). Archipelago regulates cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 413 311–316. 10.1038/35095068 [DOI] [PubMed] [Google Scholar]
- Nakamura A. J., Rao V. A., Pommier Y., Bonner W. M. (2010). The complexity of phosphorylated H2AX foci formation and DNA repair assembly at DNA double-strand breaks. Cell Cycle 9 389–397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nash P., Tang X. J., Orlicky S., Chen Q. H., Gertler F. B., Mendenhall M. D., et al. (2001). Multisite phosphorylation of a CDK inhibitor sets a threshold for the onset of DNA replication. Nature 414 514–521. 10.1038/35107009 [DOI] [PubMed] [Google Scholar]
- Nateri A. S., Riera-Sans L., Da Costa C., Behrens A. (2004). The ubiquitin ligase SCFFbw7 antagonizes apoptotic JNK signaling. Science 303 1374–1378. 10.1126/science.1092880 [DOI] [PubMed] [Google Scholar]
- Negi A., Murphy P. V. (2021). Development of Mcl-1 inhibitors for cancer therapy. Eur. J. Med. Chem. 210:113038. [DOI] [PubMed] [Google Scholar]
- Olson B. L., Hock M. B., Ekholm-Reed S., Wohlschlegel J. A., Dev K. K., Kralli A., et al. (2008). SCFCdc4 acts antagonistically to the PGC-1 alpha transcriptional coactivator by targeting it for ubiquitin-mediated proteolysis. Gene Dev. 22 252–264. 10.1101/gad.1624208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong S. E., Mann M. (2006). A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 1 2650–2660. 10.1038/nprot.2006.427 [DOI] [PubMed] [Google Scholar]
- Onoyama I., Tsunematsu R., Matsumoto A., Kimura T., de Alboran I. M., Nakayama K., et al. (2007). Conditional inactivation of Fbxw7 impairs cell-cycle exit during T cell differentiation and results in lymphomatogenesis. J. Exp. Med. 204 2875–2888. 10.1084/jem.20062299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Benavente B., Garcia J. L., Rodriguez M. S., de Mora J. F., Farras R. (2011). GSK3-SCFFbw7 targets junB for degradation in G2 to preserve chromatid cohesion. FEBS J. 278 473–474. 10.1038/onc.2012.235 [DOI] [PubMed] [Google Scholar]
- Rajagopalan H., Jallepalli P. V., Rago C., Velculescu V. E., Kinzler K. W., Vogelstein B., et al. (2004). Inactivation of hCDC4 can cause chromosomal instability. Nature 428 77–81. [DOI] [PubMed] [Google Scholar]
- Roos W. P., Thomas A. D., Kaina B. (2016). DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 16 20–33. [DOI] [PubMed] [Google Scholar]
- Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S. (2004). Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu Rev. Biochem. 73 39–85. [DOI] [PubMed] [Google Scholar]
- Schulein C., Eilers M., Popov N. (2011). PI3K-dependent phosphorylation of Fbw7 modulates substrate degradation and activity. FEBS Lett. 585 2151–2157. 10.1016/j.febslet.2011.05.036 [DOI] [PubMed] [Google Scholar]
- Sheng W., LaFleur M. W., Nguyen T. H., Chen S., Chakravarthy A., Conway J. R., et al. (2018). LSD1 ablation stimulates anti-tumor immunity and enables checkpoint blockade. Cell 174 549–563.e19. 10.1016/j.cell.2018.05.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skaar J. R., Pagan J. K., Pagano M. (2014). SCF ubiquitin ligase-targeted therapies. Nat. Rev. Drug Discov. 13 889–903. 10.1038/nrd4432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spruck C. H., Strohmaier H., Sangfelt O., Muller H. M., Hubalek M., Muller-Holzner E., et al. (2002). hCDC4 gene mutations in endometrial cancer. Cancer Res. 62 4535–4539. [PubMed] [Google Scholar]
- Srivastava M., Raghavan S. C. (2015). DNA double-strand break repair inhibitors as cancer therapeutics. Chem. Biol. 22 17–29. [DOI] [PubMed] [Google Scholar]
- Strebhardt K. (2010). Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy. Nat. Rev. Drug Discov. 9 643–U24. 10.1038/nrd3184 [DOI] [PubMed] [Google Scholar]
- Strohmaier H., Spruck C. H., Kaiser P., Won K. A., Sangfelt O., Reed S. I. (2001). Human F-box protein hCdc4 targets cyclin E for proteolysis and is mutated in a breast cancer cell line. Nature 413 316–322. 10.1038/35095076 [DOI] [PubMed] [Google Scholar]
- Sundqvist A., Bengoechea-Alonso M. T., Ye X., Lukiyanchuk V., Jin J. P., Harper J. W., et al. (2005). Control of lipid metabolism by phosphorylation-dependent degradation of the SREBP family of transcription factors by SCFFbw7. Cell Metab. 1 379–391. 10.1016/j.cmet.2005.04.010 [DOI] [PubMed] [Google Scholar]
- Suryo Rahmanto A., Savov V., Brunner A., Bolin S., Weishaupt H., Malyukova A., et al. (2016). FBW7 suppression leads to SOX9 stabilization and increased malignancy in medulloblastoma. EMBO J. 35 2192–2212. 10.15252/embj.201693889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki H., Chiba T., Suzuki T., Fujita T., Ikenoue T., Omata M., et al. (2000). Homodimer of two F-box proteins betaTrCP1 or betaTrCP2 binds to IkappaBalpha for signal-dependent ubiquitination. J. Biol. Chem. 275 2877–2884. [DOI] [PubMed] [Google Scholar]
- Tan M. J., Zhao Y. C., Kim S. J., Liu M., Jia L. J., Saunders T. L., et al. (2011). SAG/RBX2/ROC2 E3 ubiquitin ligase is essential for vascular and neural development by targeting NF1 for degradation. Dev. Cell 21 1062–1076. 10.1016/j.devcel.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Orlicky S., Lin Z., Willems A., Neculai D., Ceccarelli D., et al. (2007). Suprafacial orientation of the SCFCdc4 dimer accommodates multiple geometries for substrate ubiquitination. Cell 129 1165–1176. 10.1016/j.cell.2007.04.042 [DOI] [PubMed] [Google Scholar]
- Teng C. L., Hsieh Y. C., Phan L., Shin J., Gully C., Velazquez-Torres G., et al. (2012). FBXW7 is involved in Aurora B degradation. Cell Cycle 11 4059–4068. 10.4161/cc.22381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V., Kaur E., Kharat S. S., Hussain M., Damodaran A. P., Kulshrestha S., et al. (2019). Abrogation of FBW7 alpha-dependent p53 degradation enhances p53’s function as a tumor suppressor. J. Biol. Chem. 294 13224–13232. 10.1074/jbc.AC119.008483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsvetkov L., Stern D. F. (2005). Interaction of chromatin-associated Plk1 and Mcm7. J. Biol. Chem. 280 11943–11947. 10.1074/jbc.M413514200 [DOI] [PubMed] [Google Scholar]
- Van H. T., Santos M. A. (2018). Histone modifications and the DNA double-strand break response. Cell Cycle 17 2399–2410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Lane D., Levine A. J. (2000). Surfing the p53 network. Nature 408 307–310. [DOI] [PubMed] [Google Scholar]
- Wade M., Li Y. C., Wahl G. M. (2013). MDM2, MDMX and p53 in oncogenesis and cancer therapy. Nat. Rev. Cancer 13 83–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. M., Chan B., Tong M., Paung Y. T., Jo U., Martin D., et al. (2019). Prolyl isomerization of FAAP20 catalyzed by PIN1 regulates the Fanconi anemia pathway. PLoS Genet. 15:e1007983. 10.1371/journal.pgen.1007983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. M., Jo U., Joo S. Y., Kim H. (2016). FBW7 regulates DNA interstrand cross-link repair by modulating FAAP20 degradation. Oncotarget. 7 35724–35740. 10.18632/oncotarget.9595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L. H., Chen R. P., Li G., Wang Z. G., Liu J., Liang Y., et al. (2020). FBW7 mediates senescence and pulmonary fibrosis through telomere uncapping. Cell Metab. 32:860. 10.1016/j.cmet.2020.10.004 [DOI] [PubMed] [Google Scholar]
- Wang Q. Q., Song Y. J., Xue Y., Yu H. (2017). E3 ligase FBXW7 is critical for RIG-I stabilization during antiviral responses. Nat. Commun. 8:14654. 10.1038/ncomms14654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Wang Y., Liu N., Ren C. G., Jiang C., Zhang K., et al. (2013). FBW7 regulates endothelial functions by targeting KLF2 for ubiquitination and degradation. Cell Res. 23 803–819. 10.1038/cr.2013.42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Inuzuka H., Zhong J., Wan L., Fukushima H., Sarkar F. H., et al. (2012). Tumor suppressor functions of FBW7 in cancer development and progression. FEBS Lett. 586 1409–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei W., Jin J., Schlisio S., Harper J. W., Kaelin W. G., Jr. (2005). The v-Jun point mutation allows c-Jun to escape GSK3-dependent recognition and destruction by the Fbw7 ubiquitin ligase. Cancer Cell 8 25–33. 10.1016/j.ccr.2005.06.005 [DOI] [PubMed] [Google Scholar]
- Welcker M., Clurman B. E. (2007). Fbw7/hCDC4 dimerization regulates its substrate interactions. Cell Div. 2:7. 10.1186/1747-1028-2-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Clurman B. E. (2008). FBW7 ubiquitin ligase: a tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 8 83–93. 10.1038/nrc2290 [DOI] [PubMed] [Google Scholar]
- Welcker M., Larimore E. A., Frappier L., Clurman B. E. (2011). Nucleolar targeting of the fbw7 ubiquitin ligase by a pseudosubstrate and glycogen synthase kinase 3. Mol. Cell Biol. 31 1214–1224. 10.1128/MCB.01347-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Larimore E. A., Swanger J., Bengoechea-Alonso M. T., Grim J. E., Ericsson J., et al. (2013). Fbw7 dimerization determines the specificity and robustness of substrate degradation. Genes Dev. 27 2531–2536. 10.1101/gad.229195.113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welcker M., Orian A., Jin J. P., Grim J. A., Harper J. W., Eisenman R. N., et al. (2004). The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci. U.S.A. 101 9085–9090. 10.1073/pnas.0402770101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wertz I. E., Kusam S., Lam C., Okamoto T., Sandoval W., Anderson D. J., et al. (2011). Sensitivity to antitubulin chemotherapeutics is regulated by MCL1 and FBW7. Nature 471 110–114. 10.1038/nature09779 [DOI] [PubMed] [Google Scholar]
- Wrann S., Kaufmann M. R., Wirthner R., Stiehl D. P., Wenger R. H. (2013). HIF mediated and DNA damage independent histone H2AX phosphorylation in chronic hypoxia. Biol. Chem. 394 519–528. 10.1515/hsz-2012-0311 [DOI] [PubMed] [Google Scholar]
- Wu G. Y., Hubbard E. J. A., Kitajewski J. K., Greenwald I. (1998). Evidence for functional and physical association between Caenorhabditis elegans SEL-10, a Cdc4p-related protein, and SEL-12 presenilin. Proc. Natl. Acad. Sci. U.S.A. 95 15787–15791. 10.1073/pnas.95.26.15787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu R. C., Feng Q., Lonard D. M., O’Malley B. W. (2007). SRC-3 coactivator functional lifetime is regulated by a phospho-dependent ubiquitin time clock. Cell 129 1125–1140. 10.1016/j.cell.2007.04.039 [DOI] [PubMed] [Google Scholar]
- Wurtele H., Verreault A. (2006). Histone post-translational modifications and the response to DNA double-strand breaks. Curr. Opin. Cell Biol. 18 137–144. [DOI] [PubMed] [Google Scholar]
- Xiao D., Yue M., Su H., Ren P., Jiang J., Li F., et al. (2016). Polo-like kinase-1 regulates Myc stabilization and activates a feedforward circuit promoting tumor cell survival. Mol. Cell 64 493–506. 10.1016/j.molcel.2016.09.016 [DOI] [PubMed] [Google Scholar]
- Xie C. M., Tan M., Lin X. T., Wu D., Jiang Y., Tan Y., et al. (2019). The FBXW7-SHOC2-raptor axis controls the cross-talks between the RAS-ERK and mTORC1 signaling pathways. Cell Rep. 26 3037–3050.e4. 10.1016/j.celrep.2019.02.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., et al. (2004). Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23 2116–2125. 10.1038/sj.emboj.7600217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokobori T., Yokoyama Y., Mogi A., Endoh H., Altan B., Kosaka T., et al. (2014). FBXW7 mediates chemotherapeutic sensitivity and prognosis in NSCLCs. Mol. Cancer Res. 12 32–37. 10.1158/1541-7786.MCR-13-0341 [DOI] [PubMed] [Google Scholar]
- Yoo H. Y., Kumagai A., Shevchenko A., Shevchenko A., Dunphy W. G. (2004). Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 117 575–588. 10.1016/s0092-8674(04)00417-9 [DOI] [PubMed] [Google Scholar]
- Yumimoto K., Matsumoto M., Onoyama I., Imaizumi K., Nakayama K. I. (2013). F-box and WD repeat domain-containing-7 (Fbxw7) protein targets endoplasmic reticulum-anchored osteogenic and chondrogenic transcriptional factors for degradation. J. Biol. Chem. 288 28488–28502. 10.1074/jbc.M113.465179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zehir A., Benayed R., Shah R. H., Syed A., Middha S., Kim H. R., et al. (2017). Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 23 703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Shao F., Guo W., Gao Y., He J. (2019). Knockdown of KLF5 promotes cisplatin-induced cell apoptosis via regulating DNA damage checkpoint proteins in non-small cell lung cancer. Thorac. Cancer 10 1069–1077. 10.1111/1759-7714.13046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P., Shao Y., Quan F., Liu L., Yang J. (2021). FBP1 enhances the radiosensitivity by suppressing glycolysis via the FBXW7/mTOR axis in nasopharyngeal carcinoma cells. Life Sci. 283:119840. 10.1016/j.lfs.2021.119840 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Karnak D., Tan M., Lawrence T. S., Morgan M. A., Sun Y. (2016). FBXW7 facilitates nonhomologous end-joining via K63-linked polyubiquitylation of XRCC4. Mol. Cell 61 419–433. 10.1016/j.molcel.2015.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q., Mady A. S. A., Ma Y. Y., Ryan C., Lawrence T. S., Nikolovska-Coleska Z., et al. (2019). The WD40 domain of FBXW7 is a poly(ADP-ribose)-binding domain that mediates the early DNA damage response. Nucleic Acids Res. 47 4039–4053. 10.1093/nar/gkz058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao D., Zheng H. Q., Zhou Z. M., Chen C. S. (2010). The Fbw7 tumor suppressor targets KLF5 for ubiquitin-mediated degradation and suppresses breast cell proliferation. Cancer Res. 70 4728–4738. 10.1158/0008-5472.CAN-10-0040 [DOI] [PubMed] [Google Scholar]
- Zhao J. G., Tang J., Men W. F., Ren K. M. (2012). FBXW7-mediated degradation of CCDC6 is impaired by ATM during DNA damage response in lung cancer cells. FEBS Lett. 586 4257–4263. 10.1016/j.febslet.2012.10.029 [DOI] [PubMed] [Google Scholar]
- Zhao J. J., Xiong X. L., Li Y., Liu X., Wang T., Zhang H., et al. (2018). Hepatic F-Box protein FBXW7 maintains glucose homeostasis through degradation of fetuin-A. Diabetes 67 818–830. 10.2337/db17-1348 [DOI] [PubMed] [Google Scholar]
- Zhao X., Hirota T., Han X., Cho H., Chong L. W., Lamia K., et al. (2016). Circadian amplitude regulation via FBXW7-targeted REV-ERB alpha degradation. Cell 165 1644–1657. 10.1016/j.cell.2016.05.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. C., Sun Y. (2013). Cullin-RING ligases as attractive anti-cancer targets. Curr. Pharm. Des. 19 3215–3225. [DOI] [PMC free article] [PubMed] [Google Scholar]