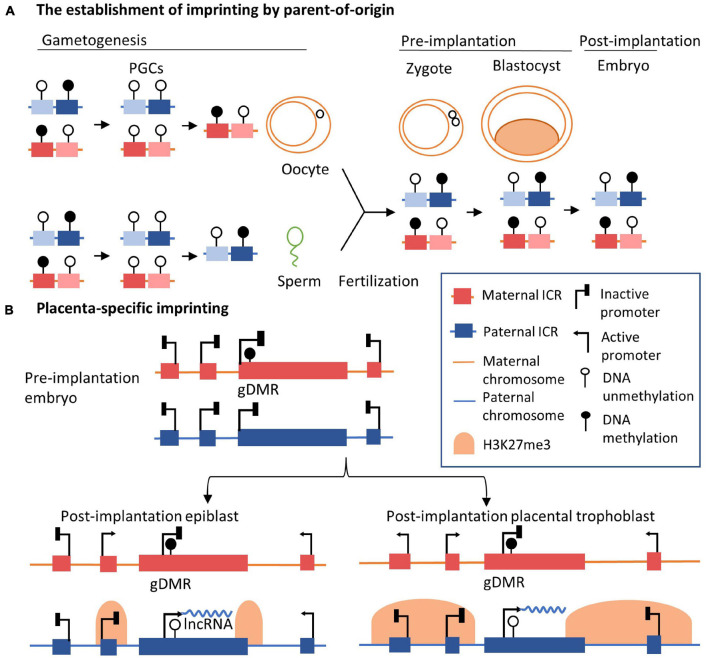
FIGURE 1.
Genomic basis of the regulation of imprinting clusters. (A) The inheritance of allele-specific imprinting epigenetic marks across generations. In the early primordial germ cells, epigenetic modifications are erased at a genomic scale before the formation of germline cells. In the germline, parent-of-origin DNA methylation is established, shown as gDMRs. After fertilization and the formation of the zygote, the gDMRs are further maintained. Established imprinting patterns are maintained in blastocyte and somatic cells in adult tissues. (B) Imprinting in epiblast and placenta in imprinting loci, such as the Kcnq1ot1/Kcnq1 or Airn/Igf2r loci, is shown. In pre-implantation embryos, DNA methylation is inherited in the gDMR on the maternal allele, such as KvDMR1 of the Kcnq1ot1/Kcnq1 imprinting cluster. After implantation, the expression of lncRNA on the maternal allele is repressed by DNA methylation in gDMR, allowing the expression of neighbor genes. On the contrary, lncRNA is expressed from the paternal allele, inducing the spreading of H3K27me3 modifications in adjacent regions in the embryonic lineage (epiblast). In extra-embryonic lineage (placental trophoblast), the extended scale of H3K27me3 marks is longer than that seen in embryonic cells. Adjacent genes further away are also silenced on the paternal allele, indicating placenta-specific imprinting, such as Slc22a18 and Tssc4 genes in the Kcnq1ot1/Kcnq1 imprinting cluster. For simplicity, specific gene names are not shown.