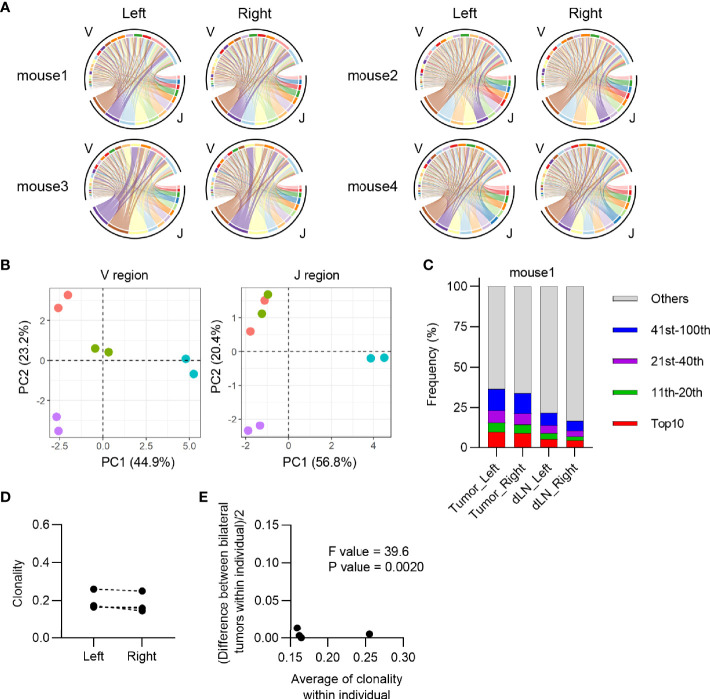
Figure 2.
Characteristics of CD4+ T cell repertoire in the bilateral tumor. (A), V/J segment usage plots of the bilateral CD4+ T cell repertoires. Ribbons connecting the V and J segments are scaled by the corresponding V/J pair frequency. The color of the circumference represents the same V and J genes, and the color of the ribbon represents the same J genes (n = 4). (B), Principal component analysis of V and J segment usage of tumor-infiltrating CD4+ T cells. Dots of the same color indicate left and right tumors of the same individual (n = 4). (C), Frequency of abundant CD4+ T cell clones in the tumor and dLN. CD4+ T cell clones were categorized into five classes based on their rank in each repertoire: top 10, 11th–20th, 21st–40th, 41st–100th, and others. The total frequency of clones in each class is shown (n = 4). (D), Clonality of the left and right CD4+ tumor repertoire (n = 4). (E), Homoscedasticity plot for variance of clonality in individual mice. The X-axis represents the average clonality of bilateral tumors in each individual (n = 4). The Y-axis represents variance of clonality between different bilateral tumors within individuals. Sum of squares within mouse = 4.4 × 10-4, Sum of squares between mice = 1.3 × 10-2, Degree of freedom within mouse = 4, Degree of freedom between mice = 3.