Figure EV2. The microRNAome of learning‐related brain regions in the aging mouse brain.
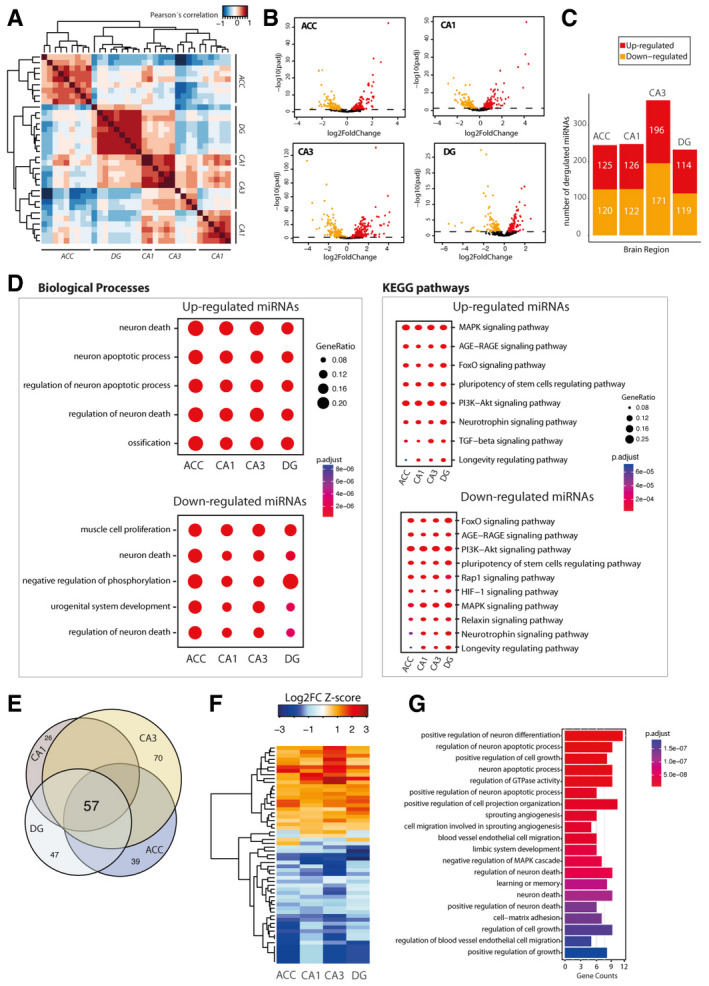
We performed small RNA sequencing of the hippocampal sub‐regions CA1, CA3, and dentate gyrus (DG) and the anterior cingulate cortex (ACC) isolated from young (3 months of age) and cognitively impaired old (16.5 months of age) mice.
-
APearson’s coefficient‐based correlation followed by unsupervised clustering of all small RNA‐seq data from different brain regions (ACC; CA1; CA3; DG) in young mice reveals brain region‐specific expression of the microRNAs that were particularly obvious for the ACC versus the hippocampal sub‐regions and the DG versus CA1 and CA3. These region‐specific differences were, however, mainly attributed to different expression values since the majority of the microRNAs, namely 176 microRNAs, could be detected at reliable levels in all investigated brain regions.
-
BVolcano plots showing differential expression of microRNAs in the different brain regions when comparing young versus old mice.
-
CBar plots showing the number of up and downregulated microRNAs in the investigated brain regions.
-
DComparison of gene ontology and functional pathways of experimentally confirmed target genes of the deregulated microRNAs in the investigated brain regions. Pathway is generally linked to neuronal death and longevity pathways.
-
EVenn diagram showing that 57 microRNAs are commonly deregulated in the aging brain.
-
FHeat map showing hierarchical clustering of the 57 commonly deregulated microRNAs based on Log2 fold change Z‐score.
-
GTop 20 biological processes affected by the confirmed target genes of the 57 commonly deregulated microRNAs in the aging brain.
