Figure 1. Repeat RNA‐tagging system enables identification of CGG repeat specific RNA‐binding proteins within cells.
-
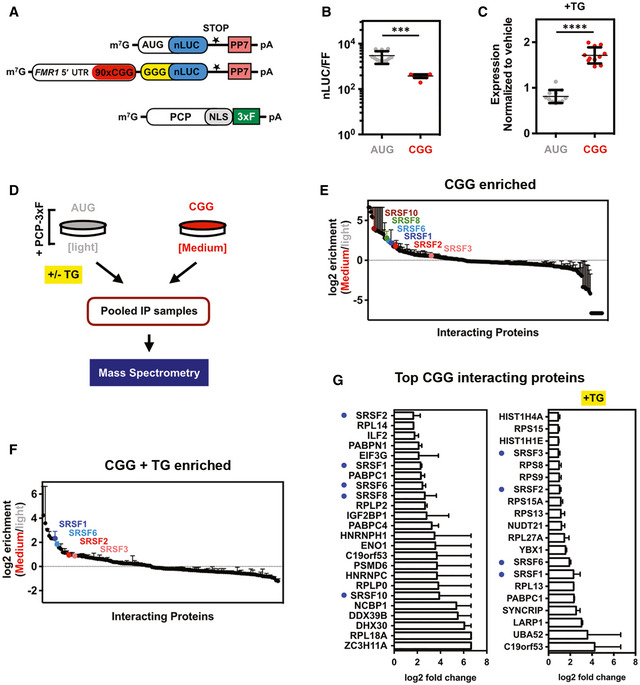
ASchematic of PP7‐tagged RNA reporters and PCP‐NLS‐FLAG constructs used in this study. Details of reporter sequences are described in Appendix Table S1.
-
BRelative protein expression from PP7‐tagged CGG‐nLuc reporters compared to AUG‐driven reporters in HEK293T cells (n = 12 biological replicates).
-
CExpression of AUG‐control and CGG‐nLuc RAN translation reporters in HEK293T cells treated with the ER stress agent thapsigargin (TG, 2 μM) (n = 11 biological replicates) normalized to vehicle (DMSO).
-
DSchematic of immunoprecipitation and mass spectrometry experiments aimed at identifying CGG repeat RNA‐interacting proteins (see Materials and Methods for details). Growth media, containing different isotopes of lysine and arginine for SILAC labeling, are indicated as light (Lys‐0 and Arg‐0) and medium (Lys‐4 and Arg‐6).
-
E, FLog2 fold‐change of the CGG‐interacting protein enrichment compared AUG reporter (n = 2 independent experiments; error bars represent range between repeats) under normal (E) and after integrated stress response activation by TG (F).
-
GList of top proteins enriched in CGG‐PP7 RNA interaction compared to AUG in mass spectrometry experiments without or with TG treatment (TG+). Bar graphs represent average of two biological replicates ± range. Enriched SR proteins are marked with blue dots.
Data information: For graphs in (B and C), error bar represents mean ± SD. Statistical analysis was performed using two‐tailed Student’s t‐test with Welch’s correction, ***P < 0.001; ****P < 0.0001.
Source data are available online for this figure.
