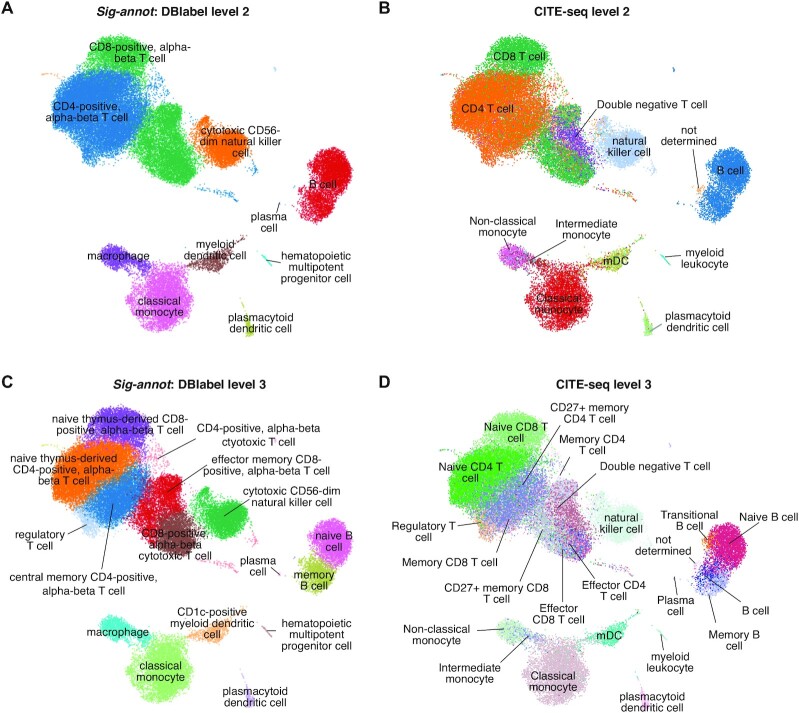
Figure 3.
Sig-annot applied to Kotliarov2020 data containing hematopoietic cells of multiple healthy donors from blood, probed by CITE-seq. (A) RNA signature-based cell type attribution at level 2, consisting of cell subtypes such as CD4+ T cells and classical monocytes. (B) Protein-marker based annotation using a gating method of classical FACS markers at a similar hierarchical depth as described in (A). Cell attribution is highly consistent with the automated RNA based results. (C, D) RNA signature-based (C) and protein-based (D) cell type attribution at the most fine-grained level 3. Even immune cell subtypes such as memory versus naive B cells or rare populations such as regulatory T cells and plasmacytoid dendritic cells are correctly attributed.