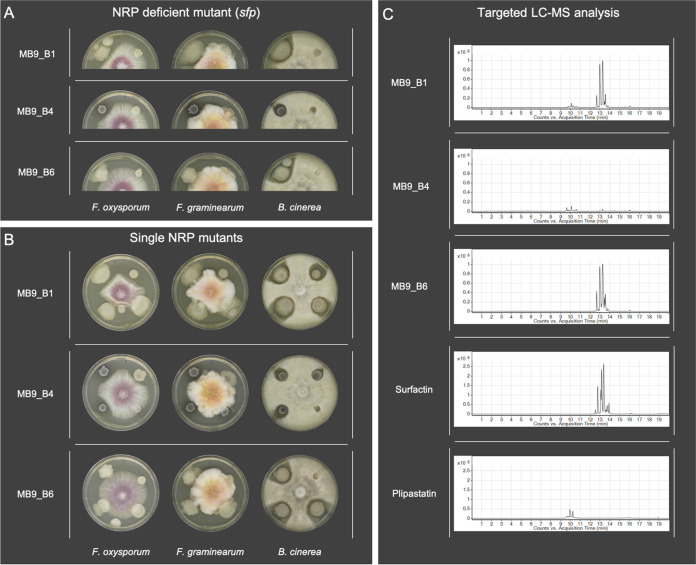
FIG 1.
(A) Antagonism assays between the plant pathogenic fungi Fusarium oxysporum, Fusarium graminearum, and Botrytis cinerea and the B. subtilis soil isolates (left) as well as their NRP-deficient sfp mutants (right). (B) Antagonism assays between the plant pathogenic fungi and B. subtilis soil isolates (upper left) as well as their single nonribosomal peptide srfAC (upper right, no surfactin), ΔppsC (lower right, no plipastatin), and ΔpksL (lower left, no bacillaene) mutants. A 5-μl quantity of bacterial overnight culture and fungal spore suspension was spotted onto the edges (bacteria) and in the center (fungi) of potato dextrose agar (PDA) plates. Strains were cocultivated at 21 to 23°C for 6 days. (C) Extracted ion chromatograms (m/z 1,000 to 1,600) display various levels of production of surfactin and plipastatin among B. subtilis soil isolates. The standard mixtures of plipastatin and surfactin are shown at the bottom. Multiple peaks in the LC-MS traces among the isolates and standards show different surfactin and plipastatin analogs with different fatty acid substitutions. The presence of surfactin and plipastatin in the isolates’ extracts was confirmed by retention time comparisons with the standards and by tandem mass spectrometry (MS/MS) fragmentation studies.