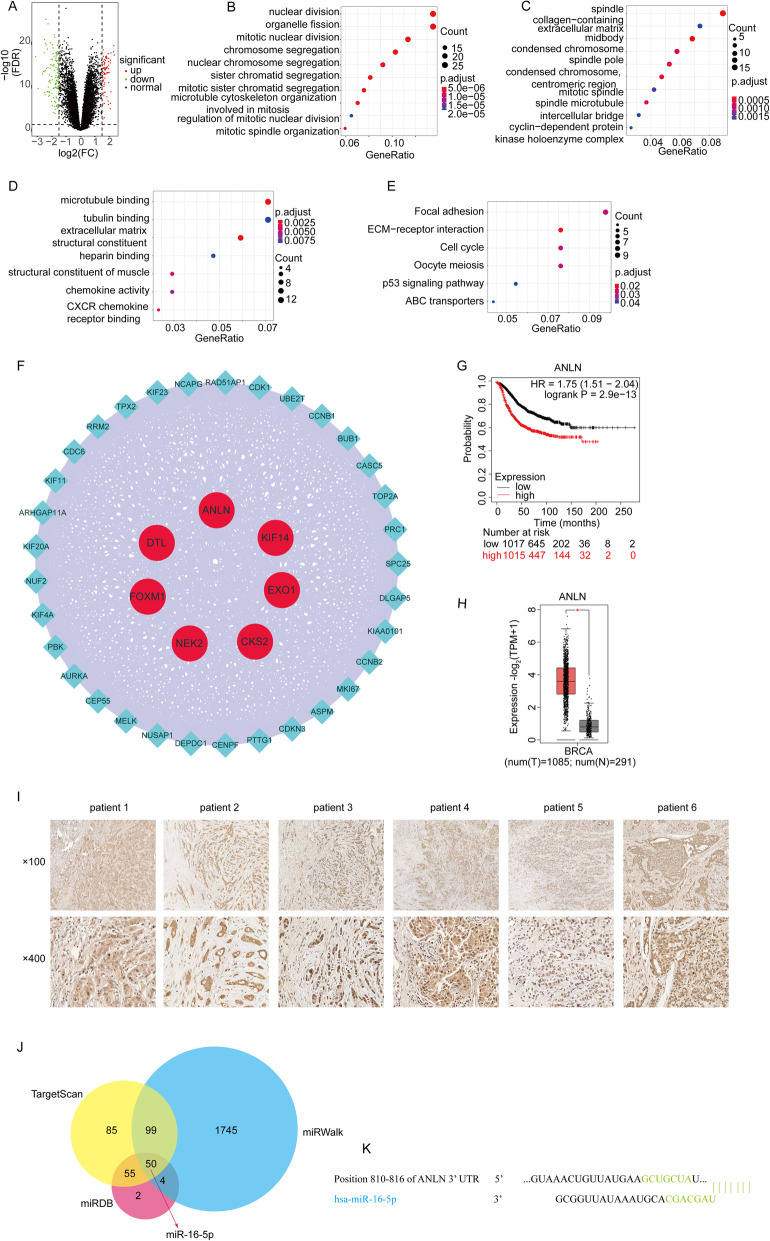
Fig. 1.
ANLN, up-regulated in BC, was a target gene of miR-16-5p. a Volcano plot of DEGs from GSE86374. b-e Dot plots of BP (b), CC (c), MF (d) and KEGG (e) enrichment analysis for DEGs. f top of module and hub genes identification from MCODE plug-in of Cytoscape software. g Survival analyses of ANLN in BC by the Kaplan Meier-plotter database. h Expression levels of ANLN in BC comparing to normal tissues. I IHC staining analysis of ANLN in 6 BC tissues. Magnification: × 100, bar = 100 μm. Magnification: × 400, bar = 20 μm. j Target miRNAs of ANLN were predicted by TargetScan, miRWalk and miRDB online databases, and the Venn diagram revealing the intersection. k The nucleotide 810–816 of ANLN 3′UTR that binding site with miR-16-5p by TargetScan. *p < 0.05. BC, breast cancer; DEGs, differentially expressed genes; IHC, Immunohistochemistry; CC, cellular component; MF, molecular function; BP, biological process; KEGG, Kyoto Encyclopedia of Genes and Genomes; FDR, false discovery rate