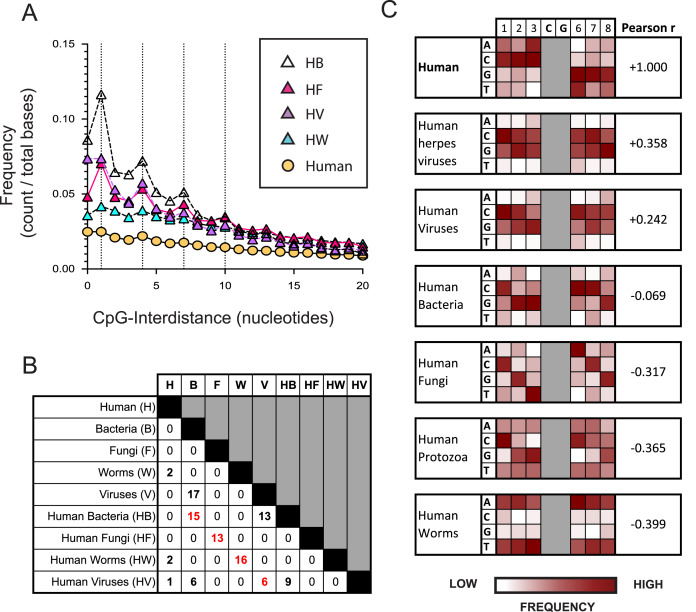
Figure 2. The nucleotide sequences flanking CpG motifs in parasitic worm genomes differ more from those in the human genome than the corresponding CpG motifs of bacteria, viruses and fungi.
(A) The distance between one CpG motif and the next in genomic datasets comprising different evolutionary groups. Datasets include the human genome (H) and genomes of the bacteria (HB), fungi (HF), viruses (HV) and parasitic worms (HW) that infect humans. Dotted vertical lines highlight the frequency of CpG interdistances of 1, 4, 7, and 10 bases. (B) The number of CpG+ octamers that are common to the twenty most frequent CpG+ octamers in each dataset. (C) Distribution of nucleotides in octamer sequences with a central CpG site in each genomic dataset. The distribution of CpG+ octamers in the human genome is compared to their distribution in each non-human dataset using the Pearson correlation coefficient.