Figure 1 .
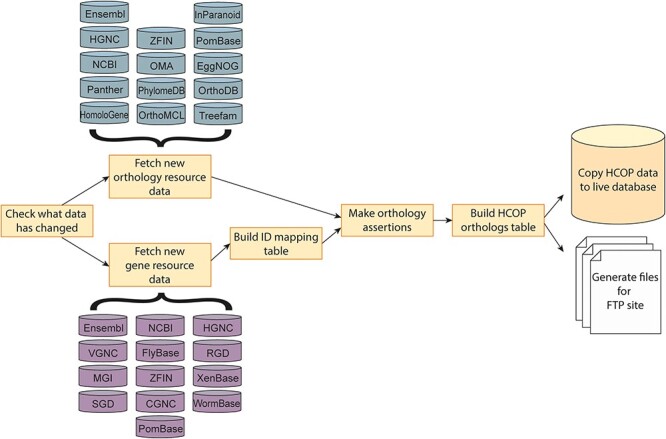
The HCOP data production pipeline. The pipeline is broken up into several stages: nomenclature/gene resource data update, orthology data update, generation of an ID mapping table which links together the MOD ID, NCBI Gene ID, Ensembl stable gene ID and UniProt identifiers for a specific gene, conversion of the raw orthology data to HCOP orthology assertions which include the appropriate gene ID information from the ID mapping table and finally combination of assertions that share the same NCBI Gene and Ensembl gene IDs to produce a single combined ortholog for each ortholog pair. Each combined ortholog is added to the HCOP orthologs table in the HCOP MySQL database, which is then used to update the public database and FTP site files.
