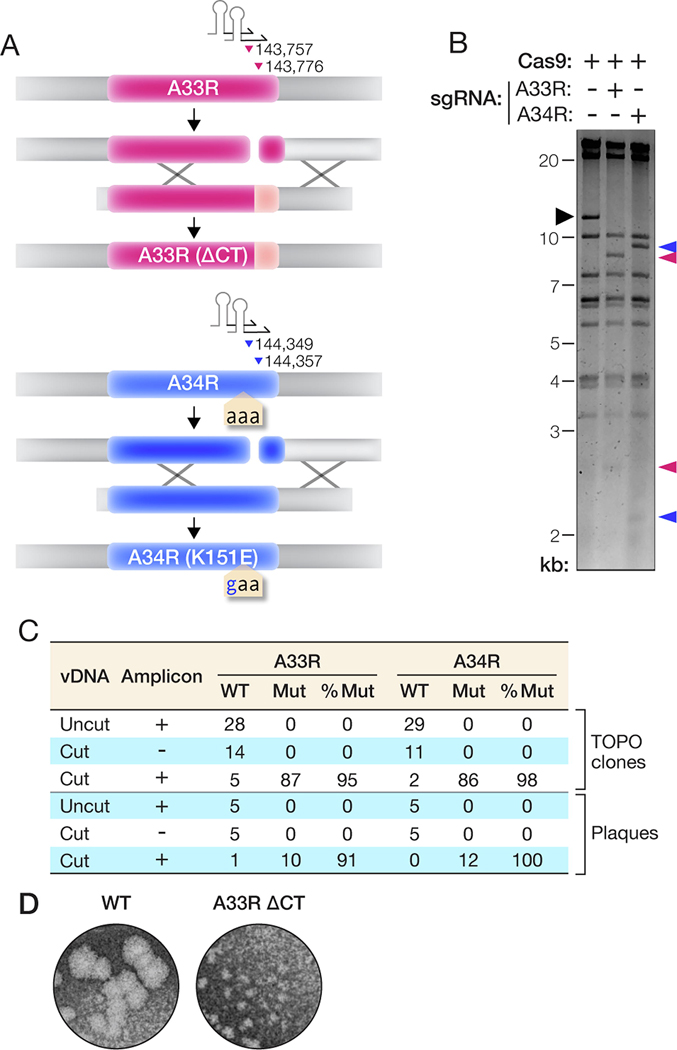
Figure 2. Mutation of A33R and A34R genes without a selectable marker.
(A) Overview of the strategy to introduce mutations into the A33R or A34R genes. The parental vDNA was cut with Cas9 directed by sgRNAs (stem-loop and arrowheads) as shown. The numbers indicate the first nucleotide of the PAM sequence for each sgRNA. The numbering is based on the VACV Western Reserve genome (NCBI RefSeq NC_006998.1). Homologous recombination (‘X’ shapes) between the transfer amplicon and cleaved vDNA promotes genome repair and insertion of mutations. (B) vDNA (incubated with Cas9 without or with sgRNAs) was fragmented by XhoI digestion, resolved by agarose gel electrophoresis, and visualized by ethidium bromide staining. Black arrowhead, vDNA fragment containing the A33R and A34R genes. Pink and blue arrowheads, new vDNA fragments observed upon cleavage with Cas9 and sgRNAs specific for A33R or A34R, respectively. (C) PCR amplicons from panel C were sequenced to determine the efficiency of rVACV rescue. The number of plasmids or PCR amplicons showing WT or mutant sequences and the corresponding percentage of mutant sequences are shown. (D) WT and A33R(ΔCT) mutant plaques were visualized by staining with crystal violet after 4 days of growth under a 0.5% methylcellulose overlay.