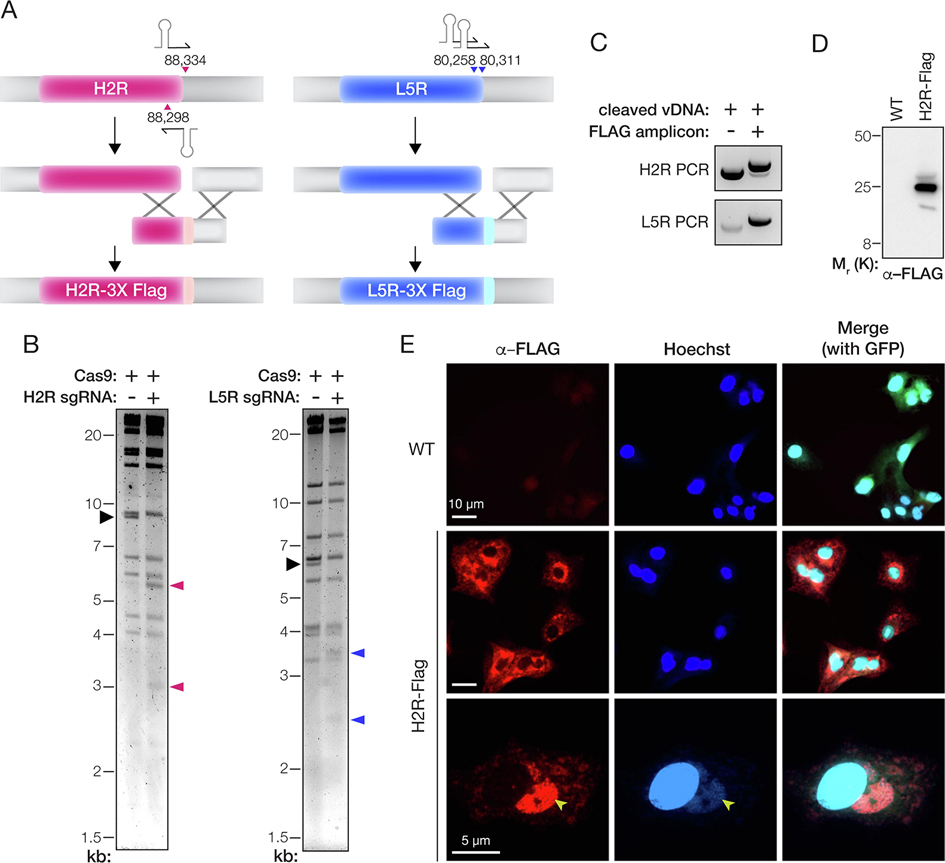
Figure 3. Epitope-tagging of genes encoding components of the entry fusion complex.
(A) Depiction of the strategy to fuse a 3X-FLAG tag to the C–terminus of the H2R and L5R proteins. The parental vDNA was cut with Cas9 directed by sgRNAs (stem-loop and arrowheads). The numbers show the first nucleotide of the PAM sequence for each sgRNA based on the VACV Western Reserve genome (NCBI RefSeq NC_006998.1). Homologous recombination (‘X’ shapes) between the transfer amplicon and cleaved vDNA promotes genome repair and insertion of the 3X-FLAG tag. (B) vDNA (incubated with Cas9 without or with sgRNAs) was fragmented by digestion with HindIII (H2R) or XhoI (L5R) digestion, resolved by agarose gel electrophoresis, and visualized by ethidium bromide staining. Black arrowhead, fragment harboring the H2R or L5R gene. Pink or blue arrowheads: vDNA fragments observed upon cleavage with Cas9 and sgRNAs specific for H2R or L5R, respectively. (C) PCR amplicons produced by amplifying the mixed population of viral genomes with gene-specific primers after rescue of transfected vDNA (without or with transfer amplicon). (D) Extracts of cells infected with WT or H2R-FLAG-tagged viruses were resolved on a polyacrylamide gel and FLAG-tagged proteins were visualized by immunoblotting. (E) Images of BSC-40 cells infected with WT or H2R- FLAG-tagged viruses and immunostained with α–FLAG antibody. Cells were counterstained with Hoechst 33342 to visualize nuclei and cytoplasmic sites of viral assembly (yellow arrowheads). eGFP expressed from the tk locus of both viruses is used as a marker for viral infection in the merge panels at the right.