Figure 6.
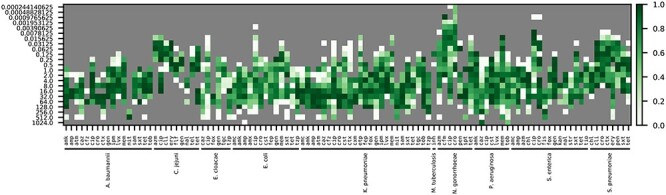
Accuracies for an XGBoost model built to predict MICs. One model was built for all species with at least two different MICs, and 450 genomes for each antibiotic using nucleotide 7-mers as input features. The coloring depicts accuracy for predicting the MIC for each antibiotic and species based on a 5-fold cross-validation. Accuracies are reported within ±1 two-fold dilution step, which is the limit of resolution for most automated MIC detection methods. Antibiotic abbreviations are defined in Table S2.
