Figure 3.
TM-score optimization and benchmark of predictions
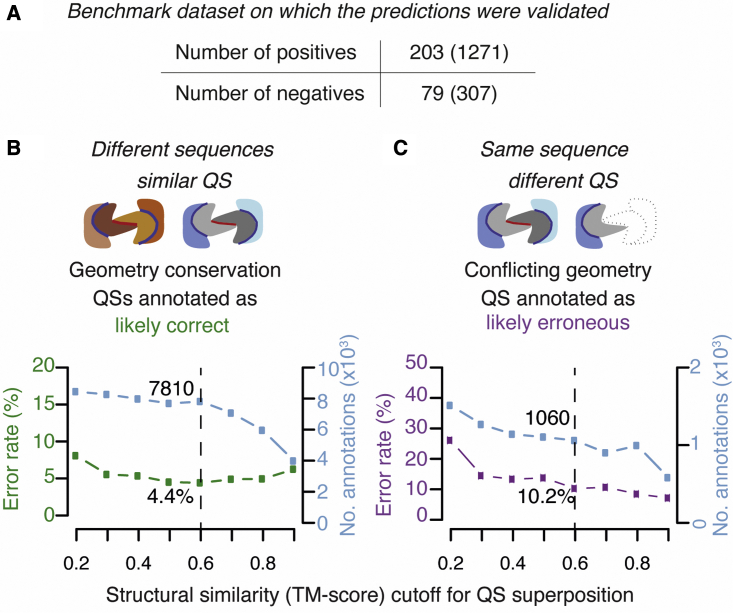
(A) Number of non-redundant structures in the manually curated benchmark dataset. Positives are correct structures and negatives are likely errors. The number of redundant structures is given in parentheses.
(B) The structural similarity score (TM-score) cut-off determines the minimum value at which two QSs are considered conserved and thereby inferred “physiologically relevant.” We scanned different TM-score cut-offs, calculated the error rate (green line), and recorded the total number of QSs annotated (blue line) for each.
(C) Starting from validated QSs, QSalignHET then searches for conflicting QSs that have identical composition and different structures (i.e., TM-score below the cut-off). We annotated such cases as likely errors and show the accuracy of these predictions (purple line) as well as the number of structures annotated for different cut-off values (light blue).