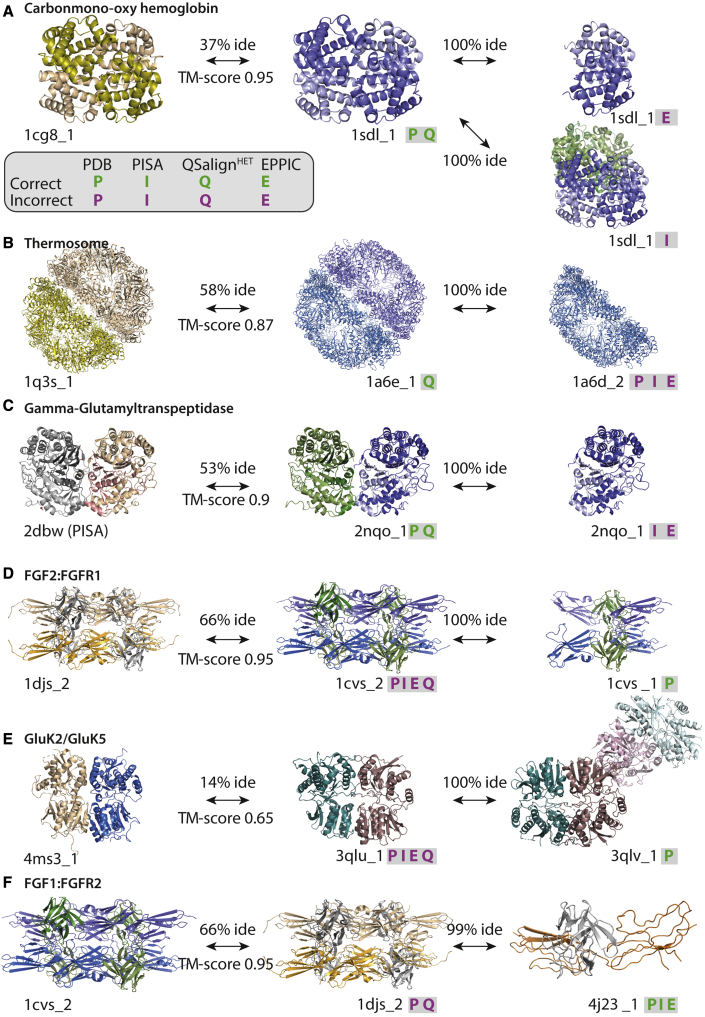
Figure 5.
Examples of prediction differences across methods
(A) EPPIC predicts human hemoglobin to be a dimer and PISA predicts it to be an octamer, whereas the conservation of the tetramer yields a correct annotation by QSalignHET.
(B) EPPIC and PISA predict half of the thermosome to be the physiologically relevant assembly, likely due to the inter-ring interface being small. However, the conservation of the two-ring structure is detected and yields a correct annotation with QSalignHET.
(C) PISA and EPPIC predict an ɑβ form for 2nqo instead of the ɑ2β2 form.
(D) The octameric structure of FGF2-FGFR1 is predicted by all methods as the correct assembly. Nevertheless, this complex is described as a tetramer in the primary reference.
(E) The dimeric structure of GluK2/GluK5 is predicted by all methods to be physiologically relevant, but the primary reference (Kumar et al., 2011) describes tetramers in solution. Based on these experimental data and considering that the tetramer's interface is observed in seven crystal forms according to PROTCID (Xu and Dunbrack, 2020), we included the tetrameric form in the PiQSiHET benchmark dataset.
(F) The dimeric structure of FGF1:FGFR2 is predicted by all the methods except QSalignHET to be physiologically relevant. However, its close homolog (99% identical) FGF2:FGFR1 has an octameric structure, and the octameric QS is found to be conserved. The octamer is observed in three crystal forms according to PROTCID.