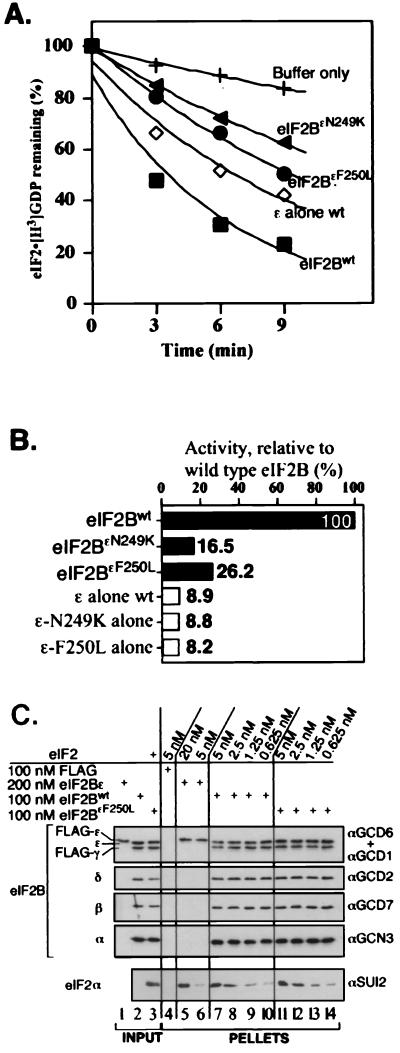
FIG. 6.
In vitro analysis of purified mutant eIF2B five-subunit complexes. (A) Nucleotide exchange assays comparing activities for mutant eIF2B complexes containing N249K (filled triangle) and F250L (filled circle) alleles of eIF2Bɛ with wild-type eIF2B (eIF2Bwt) (filled square) (1 μg each) and eIF2Bɛ alone (2.5 μg, open diamond). Assays were done in triplicate, with a standard deviation of less than 2.5 for each time point. (B) Analysis of rates of nucleotide exchange activity for mutant eIF2B complexes and isolated eIF2Bɛ subunits relative to wild-type eIF2B activity (percent initial activity). Analysis was performed as described in the legend to Fig. 2. (C) Analysis of binding between the indicated concentration of purified eIF2 and FLAG-tagged wild-type eIF2B complex (lanes 7 to 10) or eIF2BɛF250L (lanes 11 to 14). Also shown are control lanes using FLAG peptide (lane 4) or wild-type eIF2Bɛ alone (lanes 5 and 6) in place of eIF2B. Lanes 1 to 3 contain inputs; 5% of each eIF2B preparation used in the reaction mixtures was loaded and 6.25 ng of eIF2 was also loaded in lane 3 (representing 10% of 5 nM used in the reaction mixtures). Detection as described in the legend to Fig. 3, with 33% of each reaction pellet loaded to probe for eIF2 and 10% loaded to probe for each eIF2B subunit.