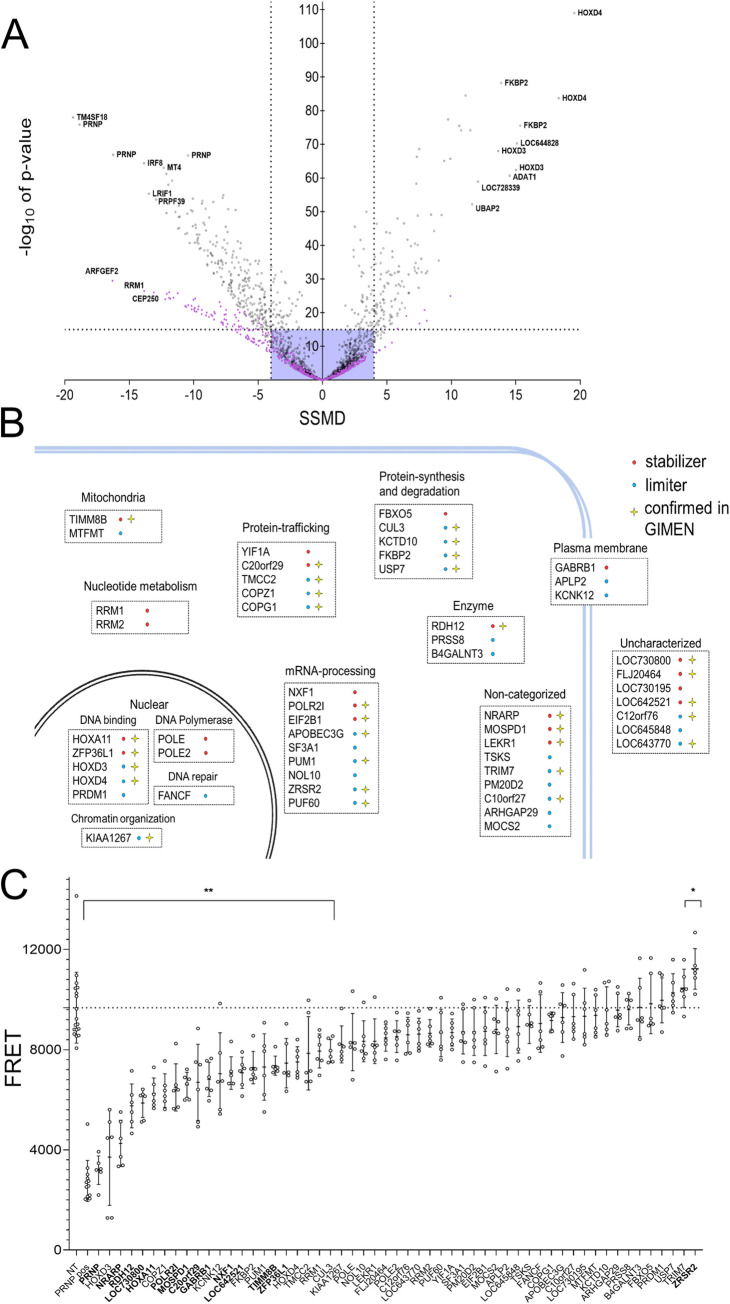
Fig 2. Secondary screening and validation of hits in smNPC-derived neurons.
(A) Extent (SSMD) and confidence (p-value) of PrPC modulation by candidate limiters (left) and stabilizers (right). Dots represent averages of triplicates; 3 siRNAs/gene were assayed. The 10 strongest limiters and stabilizers, as well as siRNAs for PRNP are highlighted. Violet box: cut-off criteria. Targets with ≥2 siRNAs with |SSMD| > 4 or p < 10−15 were considered hits (n = 54). Colors indicate screening subsets (black = first round, violet = second round). (B) Function and topology of the top 54 hits. Red: stabilizers; limiters: blue. Star: hits overlapping with GIMEN cells. (C) PrPC levels in smNPC-derived neurons transfected with a pooled set of siRNAs (as used for the primary screen, 10 nM). Dotted line: average value for cells transfected with non-target siRNAs. Genes highlighted in bold showed the same effect observed in the siRNA screening performed in U251-MG cells. n = 6 individual wells for each set of siRNAs. Values represent mean ± SD. * p = 0.0283 ** p ≥ 0.0068 (Dunnett’s multiple comparisons test).