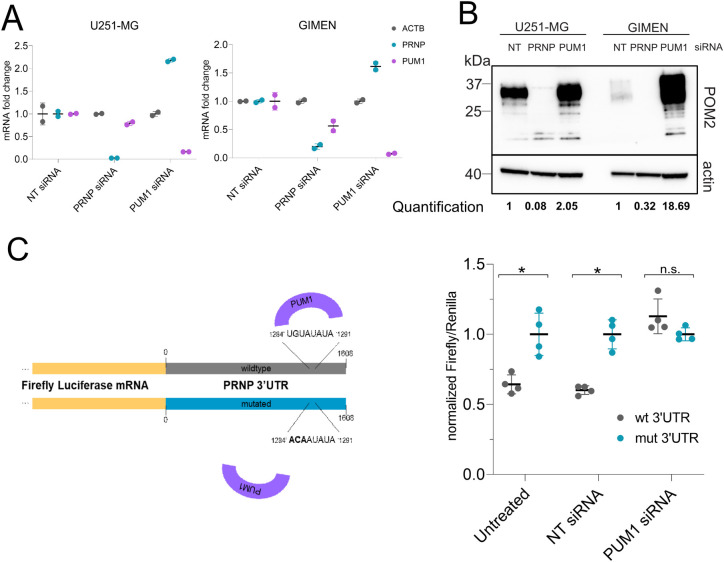
Fig 4. PUM1 regulates PRNP mRNA via its 3’UTR.
(A) qRT-PCR analysis of U251-MG and GIMEN cell lines upon transfection with PUM1 siRNA in 6-well format. PUM1 mRNA was efficiently downregulated. ΔCt values were normalized to those of β-actin (ACTB). Downregulation of PUM1 resulted in increased PRNP mRNA in both cell lines. NT (non-targeting) and PRNP siRNA were used as controls. (B) Western blot analysis of U251-MG and GIMEN cell lines upon transfection with PUM1 siRNA. PUM1 downregulation led to an upregulation of PrPC levels (as seen in Fig 3B through CRISPRi). Antibody POM2 was used for detection of PrPC. PRNP siRNA were used as controls. For quantification, signal intensity of PrPC was normalized to the signal intensity of β-actin (C) Dual-Glo Luciferase assay to assess regulation of PRNP mRNA by PUM1 via its 3’-UTR. Left panel shows a schematic of the assay. The numbers indicate the position in the 3’-UTR of PRNP. The PRNP 3’-UTR sequence, predicted to bear the consensus sequence for PUM1 binding, was placed after the gene coding for the firefly luciferase. Through a mutation in the 3’-UTR binding site of PUM1 (wt 3’-UTR = > mut 3’-UTR), the binding is prevented leading to increased expression of the firefly luciferase. Two plasmids (wt 3’-UTR and mut 3’-UTR) were co-transfected into HEK293-T cells either with none, NT, or PUM1 siRNAs. Cells transfected with the mut 3’-UTR plasmid showed a higher signal in the assay in comparison to transfection with the wt 3’UTR plasmid. Similar results were obtained for co-transfection of mut 3’-UTR plasmid and a non-targeting siRNA. The co-transfection with PUM1 siRNA abrogated the signal difference. Means ± SD (n = 4). * p ≤ 0.016, n.s. = non-significant (multiple t-test). NT: non-targeting siRNA.