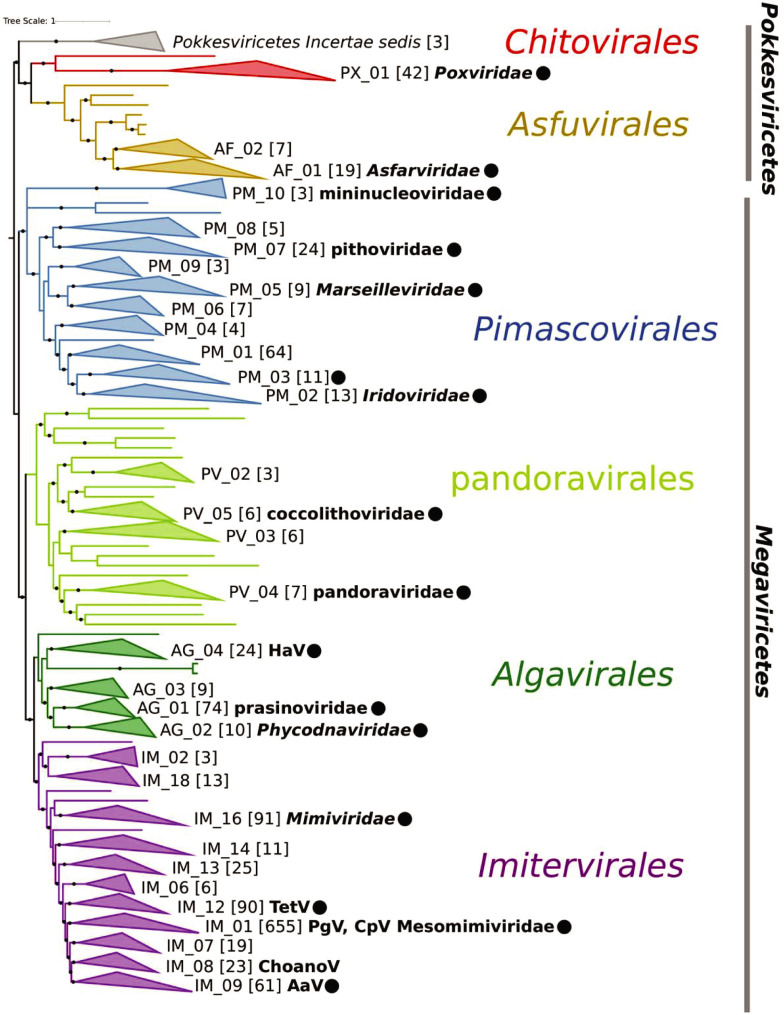
Fig 2. Phylogeny of Nucleocytoviricota based on the 7-gene marker gene set that had the highest TC value of those tested.
The phylogeny was inferred using the LG+I+F+G4 model in IQ-TREE. Solid circles denote IC values >0.5. Families are denoted by collapsed clades, with their nonredundant identifier provided at their right. The number of genomes in each clade is provided in brackets. Established family names are provided in bold italics, and proposed names are provided in lowercase. The presence of notable cultivated viruses is provided in bold next to some clades. Aav, Aureococcus anophagefferens virus; ChoanoV1, Choanoflagellate virus; CpV, Chrysochromulina parva virus; HaV, Heterosigma akashiwo virus; IC, Internode Certainty; PgV, Phaeocystis globosa virus; TC, Tree Certainty; TetV, Tetraselmis virus.