Abstract
Objectives
Assessment of whether severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has been propagated during intestinal passage and infectivity is conserved when shed rectally by hospitalized individuals.
Methods
An exploratory cohort study including 28 inpatients with coronavirus disease 2019 with estimation of RNA levels by RT-PCR and of viral infectivity by culturing of viral material sampled concomitantly and identically from pharynx and rectum.
Results
SARS-CoV-2 RNA was detected more frequently (91%, 30/33 versus 42%, 14/33, p <0.0001) and at higher concentrations (median levels 2 190 186 IU/mL versus 13 014 IU/mL, p <0.0001) in the pharyngeal swabs than in the rectal swabs. For all sample pairs (n = 33) the rectal swabs contained undetectable or lower SARS-CoV-2 RNA concentrations than their paired pharyngeal swabs. Replicative virus was found in 37% (11/30) of the PCR-positive pharyngeal swabs, whereas none of the PCR-positive rectal swabs could be cultured (0%, 0/14) despite containing SARS-CoV-2 RNA concentrations up to 1 544 691 IU/mL.
Conclusions
Our data draw into question whether SARS-CoV-2 is transmitted readily from faeces.
Keywords: Coronavirus disease 2019, Faeces, Gastrointestinal infection, Severe acute respiratory syndrome Coronavirus 2, Shedding, Transmission, Virus
Introduction
The presence of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) RNA in stool was reported early in the coronavirus disease 2019 (COVID-19) pandemic [1] and several studies have since confirmed rectal shedding of SARS-CoV-2 RNA [2]. Rectal shedding may persist for several weeks, and in some patients even surpass the period when viral RNA is detectable in respiratory samples [3,4]. These findings have led to the suggestion that SARS-CoV-2 is capable of being spread from faeces by surviving and/or propagating during its passage though the gastrointestinal tract [2,5]. The existence of such a transmission route requires that SARS-CoV-2 is still infectious when it leaves the rectum, a capacity that is demonstrated by laboratory propagation of sampled virus in cell cultures. Relatively few studies have assessed whether this capacity is conserved by the rectally shed virus, and studies so far have only analysed samples from one to five patients [[6], [7], [8], [9], [10], [11], [12]]. Of the four studies that have used reverse transcription polymerase chain reaction to unambiguously conclude on the culturing results, two reported that the virus was infectious [9,10] and the other two concluded that the sampled virus was non-infectious [8,11]. These divergent results and the lack of a systematic analysis of more patient samples has made it difficult to conclude whether SARS-CoV-2 is capable of such preservation of infectivity and hence to what extent transmission from faeces is feasible; this has recently sparked a debate on the topic [13,14]. Here, we report a systematic analysis and comparison of SARS-CoV-2 RNA concentrations and virus infectivity in sample pairs obtained from the rectum and the oropharynx of individuals with COVID-19 to assess whether rectally shed SARS-CoV-2 has propagated during gastrointestinal passage and preserved its infectious potential.
Materials and methods
The study was conducted at Odense University Hospital in Denmark, from 23 October 2020 to 17 March 2021 and was approved by the local ethics committee (ID S-20200047C). All participants signed informed consent. Individuals with COVID-19 who were admitted at the Department of Infectious Diseases with a preceding SARS-CoV-2 RT–PCR positive respiratory sample, were invited for participation. Clinical and demographic data were extracted from the patients' medical charts. The National Institutes of Health criteria were used to determine the severity of illness [15]. Upon inclusion, paired oropharyngeal and rectal swabs were collected at the same time and using the same swab type, and the patients were asked for repeated samples if they remained hospitalized for more than 2 weeks. RT-PCR and viral culturing were performed as described previously [16] and both were initiated within 4 hours after sampling. All samples were analysed on the same RT-PCR platform (Lightcycler 480 II using primers and probes for detection of the E-gene and Chemagic 360, RNA extracted with Chemagic viral DNA/RNA 300 kit H96; Perkin Elmer, Waltham, MA, USA). Crossing point (Cp) values were converted into concentrations of SARS-CoV-2 RNA in IU/mL using serial dilutions (four dilutions, 64 replicates, Cp range 24.00–36.49) of the First WHO International Standard for SARS-CoV-2 RNA (NIBSC code: 20/146, WHO International Laboratory for Biological Standards, Hertfordshire, UK). Whole-genome sequencing was used to evaluate the fraction of subgenomic RNA relative to genomic RNA in the samples (see Supplementary material, Appendix S1). Quantification of viable SARS-CoV-2 was performed in plaque assays using Vero E6 cells (ATCC CRL-1586). To further increase the chance of successfully cultivating SARS-CoV-2, aliquots were inoculated into whole Vero E6 culture flasks. Additional infection attempts were conducted in flask cultures of Caco-2 cells (ATCC HTB-37, see Supplementary material, Appendix S2). These were monitored for cytopathic effects, and supernatants were analysed by RT-PCR upon inoculation and on days 3 and 6 to detect virus proliferation and confirm virus identity. SARS-CoV-2 culturing was conducted in an approved BSL-3 laboratory (license no. 20200016905/5).
Results
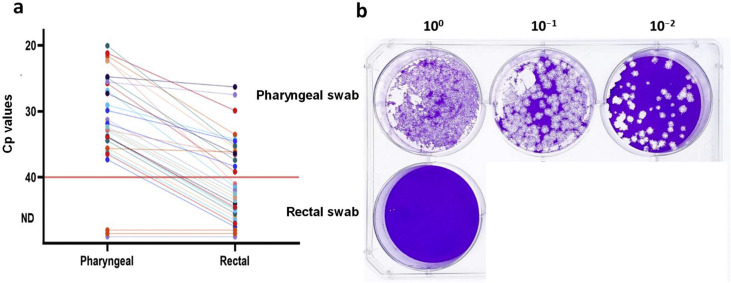
We included 28 patients in the study: 19 men and 9 women with a median age of 69 years (range 19–88 years, interquartile range (IQR) 54–77 years). Median time from onset of symptoms until sampling was 8 days (range 3–21 days, IQR 7–11 days); median time from admission until first paired sampling was 1 day (range 1–35 days, IQR 1–2 days). Upon admission, a total of 15 patients exhibited at least one GI symptom (54%): 11 patients had diarrhoea (39%) and 6 patients had nausea/vomiting (21%). Eight of the patients received proton pump inhibitors at the time of admission (29%). During hospitalization, 21 patients had severe illness (75%), three patients required intensive care (11%) and two patients died (7%). Four of the patients underwent more than one paired sampling. A total of 33 pharyngeal swabs paired with 33 rectal swabs were analysed. Thirty of the 33 (91%) pharyngeal swabs and 14 of the 33 (42%) rectal swabs were SARS-CoV-2 RNA positive by RT-PCR. This difference of 16 detected samples was statistically significant (p = 0.0001, McNemar's exact test). The median concentration of SARS-CoV-2 RNA in the RT-PCR positive rectal swabs was 13 014 IU/mL (IQR 6824–34 403 IU/mL), whereas the median concentration of SARS-CoV-2 RNA in the corresponding pharyngeal swabs was 2 190 186 IU/mL (IQR 237 929–12 446 910 IU/mL). In all cases, the concentration of SARS-CoV-2 RNA in the rectal swabs was lower than the concentration in the corresponding pharyngeal swab with a median difference of 1 711 376 IU/mL (IQR 217 724–12 434 855 IU/mL) (p <0.0001, paired sample t test; Fig. 1 a). Eleven (SARS-CoV-2 RNA concentration range 46 211–41 601 884 IU/mL) of 30 RT-PCR positive pharyngeal swabs contained culturable SARS-CoV-2 virus (37%). For three of the 11 pharyngeal swabs with culturable virus (27%) the paired rectal swab was RT-PCR negative. None of the rectal swabs, including the RT-PCR positive swabs (SARS-CoV-2 RNA concentration range 1762–1 544 691 IU/mL) contained culturable virus (Fig. 1b and Supplementary material, Figs. S1, S2). As an indicator of actively proliferating virus, the amount of subgenomic RNA relative to total RNA was measured in a subset of the pharyngeal and rectal samples but did not yield conclusive results (Supplementary material, Appendix S1).
Fig. 1.
SARS-CoV-2 RNA Cp values and infectious viral load in oropharynx and rectal swab samples from inpatients with coronavirus disease 2019. (a) Dot plot showing Cp values of SARS-CoV-2 RNA for the paired pharyngeal and rectal swabs (n = 33). Sample pairs are connected by lines. Red line indicates detection limit. (b) Representative plaque assay of paired pharyngeal and rectal swab samples in ten-fold serial dilution. No plaque-forming units were detected in the rectal swab (two wells belonging to a sample from another patient have been cropped out of the figure). Abbreviations: Cp, crossing point; ND, not detected; SARS-CoV-2, severe acute respiratory syndrome coronavirus 2.
Discussion
Propagation of rectally shed SARS-CoV-2 has been reported in five studies [6,7,9,10,12]. Three of these based their conclusion on microscopy inspections (electron microscopy of culture supernatants, inspection of cytopathic effects in inoculated cell cultures and an indirect immunoassay) [6,7,12]. Two reported the recommended confirmatory quantitative evidence for viral propagation with RT-PCR from one patient sample [10] and three patient samples [9]. In other reports, Wölfel et al. and, more recently, Albert et al. analysed thirteen and eight stool samples, respectively, and found that none could be cultured [8,11].
Here, we estimated concentrations of SARS-CoV-2 RNA and analysed for the presence of infectious virus in identically and simultaneously obtained swab samples from the rectum and the oropharynx of inpatients with COVID-19. We found that virus material obtained from these two regions differed significantly in their amount of SARS-CoV-2 RNA as well as in the infectious properties of the virus. The amount of viral RNA was consistently substantially higher in pharyngeal swabs than in the rectal swabs, with a median SARS-CoV-2 RNA concentration 168 times lower in the latter samples.
Systematic culturing showed that 37% of the pharyngeal swabs contained culturable virus, but no virus could be cultured from any of the rectal swabs in plaque assays or in highly sensitive whole-flask Vero E6 or Caco-2 cell cultures (see Supplementary material, Figs. S1, S2). This lack of viability despite relatively large amounts of viral RNA in some of the rectal swabs (up to 1 544 691 IU/mL) contrasted with the pharyngeal swabs, in which virus was culturable at concentrations down to 46 211 IU/mL.
Early in the pandemic Xiao et al. conducted histological examination and specific staining of specimens from the intestinal mucosa of individuals with COVID-19, which indicated that SARS-CoV-2 infects the intestinal epithelium [5]. The comparably much lower amounts of SARS-COV-2 RNA shed via rectum than from the oropharynx as observed in our study indicate that although the virus may infect the intestinal mucosa, it appears to not further propagate to any significant extent during gastrointestinal passage. In agreement with this, Xiao et al. reported an absence of intestinal epithelial damage in the individuals with COVID-19 [5].
In conclusion, our results from systematic culturing of 33 sample pairs obtained from individuals with COVID-19 of various severity suggests that SARS-CoV-2 is unable to survive gastrointestinal passage to any measurable extent, and if proliferation occurs in the gastrointestinal tract, the virions produced from such infection retain little to no infectivity once secreted in faeces.
Transparency declaration
The authors declare no conflicts of interest. This study was supported by the Novo Nordisk Foundation grant number NNF20SA0062931.
Authors' contributions
All authors have made a substantial contribution to the work and all participated in writing, both review and editing. Conceptualization and supervision were by RMP, TGJ, ISJ and TEA. Investigation was conducted by DST, LLB, LWM, ISJ, MNS, KS and TVS. Formal analysis was performed by RMP, DST, LWM and TEA. The original draft was written by RMP and TEA.
Editor: L. Kaiser
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.cmi.2021.10.023.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Appendix S1. Subgenomic RNA quantification from SARS-CoV-2 genomic sequence data
Appendix S2. Caco-2 culture of rectal swabs
Fig. S1. Cp values of pharyngeal and rectal swabs before and after viral culture in Vero E6 cells
Fig. S2. Cp values of rectal swabs before and after viral culture in Caco-2 cells
References
- 1.Holshue M.L., DeBolt C., Lindquist S., Lofy K.H., Wiesman J., Bruce H., et al. First case of 2019 novel coronavirus in the United States. N Engl J Med. 2020;382:929–936. doi: 10.1056/NEJMoa2001191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Guo M., Tao W., Flavell R.A., Zhu S. Potential intestinal infection and faecal–oral transmission of SARS-CoV-2. Nat Rev Gastroenterol Hepatol. 2021;18:269–283. doi: 10.1038/s41575-021-00416-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao F., Yang Y., Wang Z., Li L., Liu L., Liu Y. The time sequences of respiratory and rectal viral shedding in patients with coronavirus disease 2019. Gastroenterology. 2020;159:1158–1160. doi: 10.1053/j.gastro.2020.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Y., Guo C., Tang L., Hong Z., Zhou J., Dong X., et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–435. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xiao F., Tang M., Zheng X., Liu Y., Li X., Shan H., et al. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–1833. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xiao F., Sun J., Xu Y., Li F., Huang X., Li H., et al. Infectious SARS-CoV-2 in feces of patient with severe COVID-19. Emerg Infect Dis. 2020;26:1920–1922. doi: 10.3201/eid2608.200681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang W., Xu Y., Gao R., Lu R., Han K., Wu G., et al. Detection of SARS-CoV-2 in different types of clinical specimens. JAMA. 2020;323:1843–1844. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wölfel R., Corman V.M., Guggemos W., Seilmaier M., Zange S., Müller M.A., et al. Virological assessment of hospitalized patients with COVID-2019. Nature. 2020;581:465–469. doi: 10.1038/s41586-020-2196-x. [DOI] [PubMed] [Google Scholar]
- 9.Yao H., Lu X., Chen Q., Xu K., Chen Y., Cheng M., et al. Patient-derived SARS-CoV-2 mutations impact viral replication dynamics and infectivity in vitro and with clinical implications in vivo. Cell Discov. 2020;6:76. doi: 10.1038/s41421-020-00226-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhou J., Li C., Liu X., Chiu M.C., Zhao X., Wang D., et al. Infection of bat and human intestinal organoids by SARS-CoV-2. Nat Med. 2020;26:1077–1083. doi: 10.1038/s41591-020-0912-6. [DOI] [PubMed] [Google Scholar]
- 11.Albert S., Ruíz A., Pemán J., Salavert M., Domingo-Calap P., et al. Lack of evidence for infectious SARS-CoV-2 in feces and sewage. Eur J Clin Microbiol Infect Dis. 2021 doi: 10.1007/s10096-021-04304-4. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang Y., Chen C., Zhu S., Shu C., Wang D., Song J., et al. Isolation of 2019-nCoV from a stool specimen of a laboratory-confirmed case of the coronavirus disease 2019 (COVID-19) China CDC Wkly. 2020;2:123–124. [PMC free article] [PubMed] [Google Scholar]
- 13.Pedersen R.M., Tornby D.S., Bang L.L., Madsen L.W., Skov M.N., Jensen T.G., et al. Rectally shed SARS-CoV-2 lacks infectivity: time to rethink faecal-oral transmission? Nat Rev Gastroenterol Hepatol. 2021 doi: 10.1038/s41575-021-00501-w. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo M., Tao W., Flavell R.A., Zhu S. Reply to: Rectally shed SARS-CoV-2 lacks infectivity: time to rethink faecal–oral transmission? Nat Rev Gastroenterol Hepatol. 2021 doi: 10.1038/s41575-021-00503-8. epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.COVID-19 Treatment Guidelines Clinical Spectrum of SARS-CoV-2 Infection. https://www.covid19treatmentguidelines.nih.gov/overview/clinical-spectrum/ Available at:
- 16.Niyonkuru M., Pedersen R.M., Assing K., Andersen T.E., Skov M.N., Johansen I.S., et al. Prolonged viral shedding of SARS-CoV-2 in two immunocompromised patients, a case report. BMC Infect Dis. 2021;21:743. doi: 10.1186/s12879-021-06429-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix S1. Subgenomic RNA quantification from SARS-CoV-2 genomic sequence data
Appendix S2. Caco-2 culture of rectal swabs
Fig. S1. Cp values of pharyngeal and rectal swabs before and after viral culture in Vero E6 cells
Fig. S2. Cp values of rectal swabs before and after viral culture in Caco-2 cells