Abstract
Biliary atresia (BA) is a progressive inflammation and fibrosis of the biliary tree characterized by the obstruction of bile flow, which results in liver failure, scarring and cirrhosis. This study aimed to explore the elusive aetiology of BA by conducting whole exome sequencing for 41 children with BA and their parents (35 trios, including 1 family with 2 BA-diagnosed children and 5 child-mother cases). We exclusively identified and validated a total of 28 variants (17 X-linked, 6 de novo and 5 homozygous) in 25 candidate genes from our BA cohort. These variants were among the 10% most deleterious and had a low minor allele frequency against the employed databases: Kinh Vietnamese (KHV), GnomAD and 1000 Genome Project. Interestingly, AMER1, INVS and OCRL variants were found in unrelated probands and were first reported in a BA cohort. Liver specimens and blood samples showed identical variants, suggesting that somatic variants were unlikely to occur during morphogenesis. Consistent with earlier attempts, this study implicated genetic heterogeneity and non-Mendelian inheritance of BA.
Subject terms: Molecular biology, Genetics, Medical genetics, Mutation
Introduction
Biliary atresia is a progressive inflammation and fibrosis of the biliary tree that consequently results in the development of cholestatic liver disease. BA was first described by surgeon John Thomson in 18921 and is among the most fatal diseases, leading to severe complications in infants. The disease occurs in the early stage of neonates and can be treated by hepatic portoenterostomy or Kasai operation2. After surgical treatment, however, approximately 50% of affected infants require liver transplantation, while the rest would sustain their own liver up to the age of 5–10 years3. A study on Vietnamese BA patients reported that 84% and 71% of Kasai-treated patients survived after 1–2 years, respectively. Additionally, the respective ratios were 52% and 28% for the group without Kasai treatment4. It is estimated that after hepatic portoenterostomy operation, 70–80% of patients with BA still require liver transplantation by adulthood due to the progressive development of liver scarring, failure and cirrhosis5,6.
Although BA has been extensively studied, its aetiology and pathogenesis remain elusive. Several hypotheses explaining the cause of the disease, including viral infection, autoimmune-mediated bile duct destruction, biliary toxin, and genetic abnormality, have been proposed7. Regarding genetic aspects, debate over the Mendelian mechanism of the disease has been raised due to a lack of familial BA and a discordant presentation of BA in the monozygotic twin8. Nevertheless, some cases with familial BA have been reported, suggesting that either a recessive autosomal inheritance or a combination of genetic and acquired factors might contribute to the disease’s aetiology9–12. In addition, some studies have examined an association between BA and microchimerism, where the genetic trait is maternally transferred from the mother and later contributes to phenotypic heterogeneity and non-Mendelian inheritance13,14. More specifically, a heterozygous transition CFC1:c.433G > A in 5 BA patients with polysplenia syndrome implies a genetic predisposition to BA splenic malformation15. In a mouse model, inactivation of the hepatocyte nuclear factor 1 beta gene (Hnf1β) causes abnormalities of the gallbladder and intrahepatic bile ducts, resulting in severe jaundice16. Observations of an increased incidence of BA in some groups, such as Asian and Polynesian populations, suggest that genetic and environmental factors might cause the disease. Recent genetic studies have revealed a linkage between cholestatic jaundice and genetic predispositions in both nuclear DNA and mitochondrial DNA17–20.
The prevalence of BA is 1 in 8000–18,000 live births and varies among countries and groups, with a dominance of females over males21. The disease occurs more frequently in Southeast Asia and the Ocean Pacific22. It is approximately 1 in 5000 live births in Taiwan compared to 1 in 14,000–20,000 in North America or Western Europe6,23,24. To our knowledge, there is no epidemiological study of BA in Vietnamese, as is the prevalence of this fatal disease. The prevalence of BA in Vietnam is estimated to be as high as 1 in 2400 live births as equal to that of the Ocean Pacific regions22. Although BA and BA-related liver diseases are often observed in Vietnamese infants and are life-threatening diseases, few studies have been reported thus far4,25,26. To date, the Kasai portoenterostomy procedure has been introduced as a routine surgical practice and offers a better opportunity to patients25. However, a number of patients still require liver transplantation after the operation or have a low quality of life due to the disease’s complications. Recently, next-generation sequencing (NGS), particularly whole exome sequencing (WES), has been increasingly applied for detecting variants in patients with cholestasis27. It appears to be a powerful tool to aid diagnosis and to provide timely and accurate therapeutic treatments. Therefore, we aimed to investigate the genetic pattern of BA by conducting a family-based WES for children with BA in hope of exploring new and characterized causative variants, which would shed light on the aetiology of the deadly disease.
Materials and methods
Patient recruitment
BA diagnosis was based on intraoperative findings and liver biopsy. Patients with confirmed BA and their parents were recruited at Vinmec International Hospital and Vietnam National Hospital of Pediatrics in Hanoi from May 2019 to May 2020. A written informed consent form was provided to the parents for their participation. The study was approved by the Ethics Committee of the hospitals in accordance with the Declaration of Helsinki.
Sample collection and DNA extraction
Approximately 2 mL of peripheral blood from patients and their parents was collected in an EDTA anticoagulant tube and stored at − 80 °C. Liver wedge specimens were collected from the Kasai operation, snapped frozen in liquid nitrogen and stored at − 80 °C. Genomic DNA was extracted by using a DNA Mini Blood Isolation Kit based on the manufacturer’s protocol (Qiagen, Germany). DNA samples were quantified by fluorescence using a Qubit BR Quantification Kit (Invitrogen, USA). Extracted DNA samples were preserved at − 80 °C for future uses.
Whole exome sequencing
Exome sequencing libraries were prepared by using a Nextera Rapid Capture Kit (Illumina, Calif, USA) based on the manufacturers’ protocol with slight modifications. The library concentration was quantified by a Qubit dsDNA Broad Range Assay Kit (Invitrogen, USA). Library size was measured by using a LabChip 3 K Hisense Kit (PerkinElmer, USA). Paired-end exome sequencing with 150 bp cycles was performed on a HiSeq 4000 (Illumina, Calif, USA), targeting an averaged depth of 100X.
Bioinformatics analysis
Variant calling and annotation were performed based on highly regarded tools28. Reads with low quality, adapters and noise were removed prior to the downstream analysis by using FastQC and Trimmomatic. Reads were aligned with the reference genome GRch38 version29. Bowtie2, BWA and Qualimap were used for quality control30. To minimize the false-positive rate, multiple variant calling tools, including the Genome Analysis Toolkit (GATK)31, SAMtools mpileup32 and Freebayes33, were mutually used.
Variant classification, functional prediction and genotype–phenotype analysis
A stringent strategy was applied for variant classification, including (i) inclusion of rare and nonsynonymous variants with a minor allele frequency (MAF) < 1% against three databases: the Kinh Vietnamese population (KHV) obtained from our previous study on the Vietnamese human genome database34, GnomAD (https://gnomad.broadinstitute.org/) and 1000 Genome Project29; (ii) inclusion of variants with 3 types of inheritance modes: X-linked, homozygous and putative de novo; and (iii) variants with a CADD Phred score of > 10, indicating the 10% most deleterious variants in the genome35. In silico prediction tools, including SIFT36, PolyPhen-237, Mutation Taster38, I-Mutant39 and HOPE40, were employed to predict the impact of genetic changes. Molecular Signatures Databases (MSigDB v7.2) were used to compute the candidate genes with the gene sets of human phenotype ontology41,42.
Validation of WES results
Selected variants were then confirmed by bidirectional Sanger sequencing. Proper primers were designed for these variants, followed by PCR amplification and sequencing on an ABI 3500 DX system using a BigDye Terminator v3.1 (Thermo Fisher, Massachusetts, USA).
Ethics approval
The study was approved by the Ethics Committee of Vinmec International General Hospital JSC (Decision No. 48/2019/QD-VMEC).
Consent to participate
A written informed consent form was provided to the parents prior to their participation.
Consent for publication
The participants provided consent for publication of all relevant data and this manuscript.
Results
Clinical features
We recruited a total of 42 children who had been diagnosed with BA based on intraoperative findings and liver biopsy. All patients showed typical BA symptoms, such as prolonged jaundice, acholic stool and abnormalities of the biliary tract at early infantile. The patients, including 23 males and 19 females born from 2009 to 2019, but the majority of patients were born in recent years. All patients underwent Kasai surgery immediately after birth (mostly after their 2 months of life), but the concentrations of bilirubin and serum enzymes indicating liver function, such as ALP, ALT, AST and γ-GT, remained high at the time of enrolment (Table 1). Some patients have developed liver cirrhosis (BA002_3, BA005, BA012, BA013 and BA018). One BA patient has infected with CMV (BA037). Several probands whose siblings were reported to develop liver diseases or other genetic conditions, including BA (BA002_4), primary sclerosing cholangitis (BA032), choledochal cyst (BA042) and haemophilia (BA025). Four mothers experienced abnormal pregnancy (BA024, BA027; BA036, BA038). The remaining families did not show any significant concern during their pregnancy and had no family history of BA or other genetic conditions. Excluding one family who failed to come for blood drawing after the first health examination, we were finally able to collect blood samples from 41 BA-affected children and their parents. Among these 41 children, we collected liver specimens from 18 children obtained from the Kasai operation.
Table 1.
Clinical features of children with biliary atresia.
| Proband | Birth year | Sex | Age diagnosed | Blood test (at the time of enrolment) | Family history, clinical description | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALP (124–341 IU/L) | ALB (36–50 g/L) | ALT (< 50 IU/L) | AST (< 50 IU/L) | γ-GT (12–123 IU/L) | T-Bil (2–20 µmol/L) | D-Bil (< 8.6 µmol/L) | |||||
| BA001 | 2016 | F | 1 m/o | N/A | N/A | 190.5 | 211.9 | 30.9 | 73.59 | 36.8 | 1st child; No family history of BA or other genetic disease |
| BA002_3 | 2014 | F | 1.5 m/o | N/A | 34 | 57.8 | 109.9 | 168.4 | 143.8 | 83 | Her younger brother was diagnosed with BA; Currently, she developed signs of cirrhosis |
| BA002_4 | 2018 | M | 2 m/o | 439 | 44.8 | 163.3 | 205.4 | 1212.4 | 289.6 | 150.8 | His sister (BA002_3) showed similar CJ symptoms and diagnosed with BA |
| BA003 | 2018 | F | 50 days | N/A | N/A | 259.3 | 289.9 | N/A | 210.7 | 111.5 | 2nd child; Her grandfather's daughter died at 1 m/o and showed pale stool |
| BA004 | 2010 | M | 2 m/o | 677 | 43.3 | 282.5 | 301.5 | 820 | 20 | 5.4 | 3rd child; No family history of BA or other genetic disease; Splenomegaly; stool with fresh blood |
| BA005 | 2011 | M | 45 days | 501 | 36.8 | 153.3 | 166.7 | 243.3 | 15.7 | 4.6 | 2nd child, full term, born via C-section with birthweight of 3.5 kg; No family history of BA or other genetic disease; Developed cirrhosis after Kasai operation |
| BA006 | 2015 | M | 2 m/o | N/A | 37.1 | 107.3 | 206.8 | 200 | 132.7 | 73.3 | 1st child; No family history of BA or other genetic disease |
| BA007 | 2017 | M | 2 m/o | 275 | 41.1 | 66.1 | 85.8 | 58.8 | 8.8 | 2.2 | 1st child; No family history of BA or other genetic disease |
| BA009 | 2018 | M | 1.5 m/o | 777.2 | 30.7 | 167.6 | 249.7 | 410.5 | 238.1 | 131.7 | 3rd child; No family history of BA or other genetic disease |
| BA010 | 2010 | F | 1 m/o | 668 | 40.1 | 173.4 | 129.2 | 249.9 | 25.7 | 5.9 | 1st child, full term, C-section delivered with birthweight of 3.4 kg; No family history of BA or other genetic disease |
| BA011 | 2012 | M | 1 m/o | 310 | 45 | 50 | 67.2 | 66.2 | 7.9 | 1.3 | 2nd child; Vaginal delivered; No family history of BA or other genetic disease |
| BA012 | 2015 | F | 2.5 m/o | 748 | 41.2 | 876 | 585.2 | 624.4 | 58.2 | 30.6 | 2nd child; No family history of BA or other genetic disease. Cirrhosis developed; Splenomegaly |
| BA013 | 2010 | F | 1 m/o | 249 | 40 | 38.2 | 52.6 | 57.3 | 21.8 | 4.3 | 1st child; No family history of BA or other genetic disease; Cirrhosis developed |
| BA014 | 2016 | F | 2 m/o | N/A | 42.2 | 109.5 | 87.8 | 201.7 | 8.9 | 2.2 | 1st child; C-section delivered with birthweight of 3.4 kg; No family history of BA or other genetic disease |
| BA015 | 2016 | F | 1 m/o | N/A | 39.5 | 167.8 | 115.8 | 460 | 80.3 | 46.4 | 2nd child; No family history of BA or other genetic disease |
| BA016 | 2014 | M | 3 m/o | 353.8 | 37.09 | 110.4 | 196.1 | 424.1 | 16.8 | 5.7 | 1st child; No family history of BA or other genetic disease |
| BA017 | 2016 | F | 2 m/o | 516 | 34.6 | 161.5 | 282.7 | 224.6 | 56.8 | 26.8 | 2nd child; No family history of BA or other genetic disease |
| BA018 | 2015 | M | 2 m/o | 33.5 | 82.9 | 191.2 | 371 | 180.8 | 107.9 | 2nd child; No family history of BA or other genetic disease; Prolonged jaundice, acholic stool; cirrhosis after Kasai operation | |
| BA019 | 2017 | F | 3 m/o | 1195 | 29.7 | 64.9 | 150.9 | 384.1 | 38.8 | 16.9 | 1st child; No family history of BA or other genetic disease |
| BA020 | 2009 | M | 2 m/o | 386 | 39.8 | 87.4 | 80.7 | 176.7 | 16.7 | 5.9 | 1st child; No family history of BA or other genetic disease |
| BA021 | 2018 | F | 65 days | 584.8 | 37.6 | 220.9 | 323.1 | 918.9 | 224.1 | 123 | 1st child; No family history of BA or other genetic disease |
| BA023 | 2018 | M | 3 m/o | 635.7 | 36.75 | 163.9 | 258.5 | 404 | 153.9 | 85 | 2nd child; No family history of BA or other genetic disease |
| BA024 | 2017 | F | 2 m/o | 280.3 | 33.2 | 63.9 | 66.9 | 88 | 14.7 | 5 | A child from 2nd pregnancy; C-section delivered, full term; 1st pregnancy was a boy, stillbirth at 5 m/o of gestation due to a low level of amniotic fluid. No family history of BA or other genetic disease |
| BA025 | 2018 | F | 3 m/o | 300 | 41.2 | 175.8 | 226.3 | 465.1 | 131.1 | 82.3 | 3nd child; her older brother was with haemophilia; her older sister was healthy |
| BA026 | 2018 | M | 2 m/o | 498 | 43.2 | 178.7 | 240.5 | 781 | 76.7 | 52 | 1st child; No family history of BA or other genetic disease |
| BA027 | 2018 | M | 40 days | 497 | 38.5 | 78 | 104.9 | 565.1 | 11.3 | 4.1 | He was a child from his mother’s 3rd pregnancy; the 1st pregnancy was stillbirth at 7 weeks of gestation due to no heartbeat; the 2nd was a molar pregnancy discovered at 8 weeks of gestation |
| BA028 | 2016 | M | 28 days | 421 | 36.5 | 63.7 | 80.2 | 171.6 | 9.2 | 2.4 | 1st child of healthy parents. His paternal grandfather developed liver cirrhosis at age of 50 |
| BA029 | 2014 | M | 1 m/o | N/A | 39.5 | 221.6 | 227.8 | 527.7 | 89.9 | 51.7 | 1st child; Full term, C-section delivered with birthweight of 3.2 kg; No family history of BA or other genetic disease. Prolonged jaundice, acholic stool |
| BA030 | 2018 | M | 1 m/o | 404 | 31.4 | 123.6 | 210.2 | 900.3 | 208 | 120.4 | 2nd child; No family history of BA or other genetic disease |
| BA031 | 2018 | M | 15 days | 556 | 35.8 | 56.9 | 142.6 | 855.5 | 143.7 | 80.3 | 2nd child; No family history of BA or other genetic disease |
| BA032 | 2018 | M | 1 m/o | 427.4 | 34.4 | 76.4 | 144.6 | 144.6 | 150.6 | 69.6 | He was the 2nd child. The first child was diagnosed with primary sclerosing cholangitis and died at 28 m/o |
| BA033 | 2018 | F | 2.5 m/o | 648 | 32.8 | 134.7 | 255.2 | 131.8 | 368.8 | 188 | 2nd child; No family history of BA or other genetic disease |
| BA034 | 2018 | F | 29 days | 35.2 | 44.2 | 150.8 | N/A | 104.9 | 60.9 | 2nd child; No family history of BA or other genetic disease | |
| BA035 | 2015 | F | 72 days | 374 | 33.1 | 82.5 | 173.3 | 70 | 278.8 | 142 | 1st child; No family history of BA or other genetic disease |
| BA036 | 2018 | F | 1.5 m/o | 311 | 39.7 | 164.6 | 265.9 | 280.4 | 131.3 | 97.8 | She was a child from her mother's 2nd pregnancy. The first pregnancy was miscarriage |
| BA037 | 2018 | M | 1 m/o | 808.3 | 38.9 | 246.7 | 317.8 | 329.5 | 139.8 | 85.8 | 1st child; He was infected with CMV. His father was infected with HBV. No family history of BA or other genetic disease |
| BA038 | 2018 | M | 66 days | 240 | 39.8 | 113.8 | 87.2 | 833 | 81.4 | 47.3 | Full term, vaginal delivered with birthweight of 3.1 kg. He was a child from his mother's 3rd pregnancy; The 1st and 2nd pregnancy were stillbirth at 8 weeks of gestation. He had an inguinal hernia |
| BA039 | 2019 | F | 40 days | 618 | 36.2 | 115.4 | 130.7 | 899.3 | 130.3 | 101.2 | She was a child of her mother’s 2nd pregnancy; the first was aborted. Her maternal grandfather was with hepatitis |
| BA040 | 2019 | M | 2 m/o | 629.2 | 37.7 | 93.4 | 220 | 604.1 | 256.7 | 137 | 1st child; No family history of BA or other genetic disease |
| BA041 | 2019 | M | 28 days | 320 | 42.6 | 138.6 | 265.5 | 905.2 | 108 | 82.7 | 1st child; No family history of BA or other genetic disease. His prenatal grandmother was infected with HBV |
| BA042 | 2019 | F | 1 m/o | N/A | N/A | 211 | 663 | N/A | 229 | 120 | 2nd child; the first child was a healthy boy. Her mother was diagnosed with choledochal cyst at age of 13 |
M male, F female, m/o month old, ALP alkaline phosphatase, ALB albumin, ALT alanine aminotransferase, AST aspartate aminotransferase, γ-GT gamma-glutamyl transferase, T-Bil total bilirubin, D-Bil direct bilirubin, HBV hepatitis B, CMV cytomegalovirus, CJ cholestatic jaundice, N/A not available, BA002_3 and BA002_4 were siblings.
Genetic properties
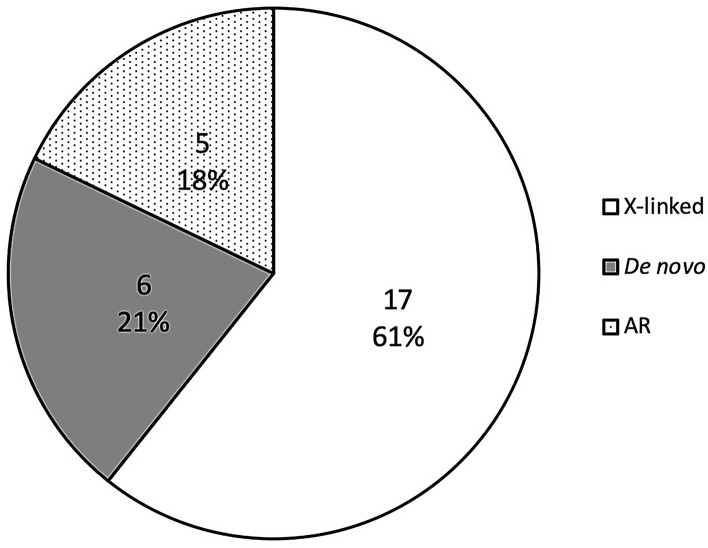
We applied a strict filtering strategy by removing variants with MAF > 1%, synonymous variants and variants with a CADD scaled score < 10. Finally, we identified a total of 28 variants in 25 genes from our BA-affected cohort (Table 2). All variants were subsequently confirmed by Sanger sequencing (Fig. S1). Among the 28 detected variants, 17 X-linked variants (61%) were detected in 17 different genes, 6 de novo variants (21%) were detected in 6 genes from 5 probands, including INVS, ELP2, TINAG, CEP63, CCDC136, and BCAR1, and 5 homozygous variants were identified in 5 genes (18%) (Fig. 1), including HACE1, VPS13C, RAPGEF4, FOCAD and INVS (Table 2). Family #2 involved two siblings with similar phenotypes (early onset jaundice, BA diagnosed). Two X-linked and 1 homozygous variants were detected in the male sib of family #2, and none were detected in his sister (Table 2). Interestingly, several genes with genetic predisposition were observed in unrelated patients, including AMER1 (BA004 and BA007), INVS (BA014 and BA041), and OCRL (BA032 and BA041). Noticeably, proband BA014 carried an INVS de novo variant, while proband BA041 carried an INVS homozygous variant (Table 2).
Table 2.
Genetic characteristics of Vietnamese children with biliary atresia.
| Proband | Sex (M/F) | Chr | Position | Gene | DNA change | MOI | Genotype | MAF (KHV/GnomAD East Asian/1 KG) | CADD (Phred) | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proband | UM | UF | AS | Allele frequency | #homozygotes | ||||||||
| BA002_4 | M | chr6 | 104,771,988 | HACE1 | NM_001350555:c.G1660 > A | AR | T/T | C/T | C/T | C/T | 0.01/0.001731/0.001996 | 0 | 26.2 |
| chrX | 72,684,557 | PHKA1 | NM_001122670:c.G478 > A | X-linked | T | C/T | C | C/T | 0.008/0.001949/0.005298 | 0 | 23.2 | ||
| chrX | 123,665,767 | THOC2 | NM_001081550:c.G1261 > A | X-linked | T | C/T | C | C/T | 0.008/0.01483/0.01589 | 0 | 21.6 | ||
| BA004 | M | chrX | 123,888,703 | XIAP | NM_001167:c.C962 > G | X-linked | G | G/C | N/A | 0/0.003356/0.003958 | 0 | 29.5 | |
| BA007 | M | chr15 | 61,929,659 | VPS13C | NM_017684:c.C5999 > G | AR | C/C | G/C | G/C | 0.01/0.005391/0.00998 | 0 | 24.3 | |
| chrX | 64,192,212 | AMER1 | NM_152424:c.A1075 > T | X-linked | A | T/A | T | 0/0.0002802/0 | 0 | 25 | |||
| chrX | 77,508,398 | ATRX | NM_138270:c.C7318 > G | X-linked | C | G/C | G | 0/0.0005613/0.001323 | 0 | 22.3 | |||
| chrX | 85,367,724 | POF1B | NM_001307940:c.A325 > C | X-linked | G | T/G | T | 0.003/0.001392/0.001321 | 0 | 19.47 | |||
| BA009 | M | chrX | 130,015,441 | BCORL1 | NM_001184772:c.G2669 > A | X-linked | A | G/A | G | 0.008/0.001949/0.002635 | 0 | 23.3 | |
| BA014 | F | chr9 | 100,252,390 | INVS | NM_001318382:c.C208 > T | De novo | C/T | C/C | N/A | 0/0/0 | 0 | 37 | |
| chrX | 40,073,898 | BCOR | NM_001123383:c.C1448 > T | X-linked | A/A | G/A | N/A | 0.003/0.003068/0.001321 | 0 | 23.1 | |||
| BA016 | M | chrX | 56,565,305 | UBQLN2 | NM_013444:c.C1432 > G | X-linked | G | C/G | C | 0/0/0 | 0 | 23.7 | |
| BA020 | M | chrX | 43,693,330 | MAOA | NM_000240:c.G208 > A | X-linked | A | G/A | G | 0/0/0 | 0 | 22.6 | |
| chrX | 108,733,510 | IRS4 | NM_003604:c.G2835 > C | X-linked | G | C/G | C | 0.003/0/0 | 0 | 22.8 | |||
| BA028 | M | chr18 | 36,156,524 | ELP2 | NM_001242879:c.C1124 > T | De novo | C/T | C/C | C/C | 0/0/0 | 0 | 26.3 | |
| BA032 | M | chr2 | 173,016,403 | RAPGEF4 | NM_001282901:c.C1204 > A | AR | A/A | C/A | C/A | 0/0.0001925/0 | 0 | 23.2 | |
| chrX | 129,590,191 | OCRL | NM_001587:c.T2603 > A | X-linked | A | T/A | T | 0.003/0/0 | 0 | 18.03 | |||
| BA033 | F | chr6 | 54,308,777 | TINAG | NM_014464:c.C227 > T | De novo | C/T | C/C | C | 0/0/0 | 0 | 22.2 | |
| BA035 | F | chr3 | 134,561,515 | CEP63 | NM_001042383:c.C1468 > A | De novo | C/A | C/C | C/C | 0/0/0 | 0 | 23.5 | |
| chr7 | 128,812,751 | CCDC136 | NM_022742:c.C2585 > A | De novo | C/A | C/C | C/C | 0/0/0 | 0 | 14.62 | |||
| BA036 | F | chr16 | 75,243,074 | BCAR1 | NM_001170715:c.C83 > T | De novo | G/A | G/G | G/G | 0/0/0 | 0 | 29.4 | |
| BA037 | M | chr9 | 20,948,857 | FOCAD | NM_017794:c.C3805 > A | AR | A/A | C/A | C/A | 0.005/0.0005763/0.001996 | 0 | 23.2 | |
| chrX | 70,341,839 | KIF4A | NM_012310:c.A1174 > C | X-linked | C | A/C | A | 0/0.005278/0.001379 | 0 | 25.1 | |||
| BA038 | M | chrX | 47,448,875 | ZNF41 | NM_001324139:c.C637 > T | X-linked | A | G/A | G | 0.003/0/0 | 0 | 15.37 | |
| BA040 | M | chrX | 3,112,294 | ARSF | NM_001201538:c.C1511 > T | X-linked | T | C/T | C | 0.003/0/0 | 0 | 22.8 | |
| chrX | 64,191,164 | AMER1 | NM_152424:c.C2123 > A | X-linked | T | G/T | G | 0.003/0/0 | 0 | 14.16 | |||
| BA041 | M | chr9 | 100,126,394 | INVS | NM_014425:c.C118 > G | AR | G/G | C/G | C/G | 0.007/0.004996/0.005988 | 0 | 22.7 | |
| chrX | 129,557,351 | OCRL | NM_000276:c.G265 > C | X-linked | C/C | G/C | G | 0.007/0.0008396/0.002639 | 0 | 24.1 | |||
Chr chromosome, M male, F female, A.A amino acid, MOI mode of inheritance, UM unaffected mother, UF unaffected father, AS affected sibling, N/A not available, MAF minor allele frequency, KHV Kinh Vietnamese, 1 KG 1000 Genome Project, CADD scaled score.
Figure 1.
Mode of inheritance of identified variants from the biliary atresia cohort. X-linked variants are presented in blank; de novo variants are presented in grey; and autosomal recessive variants are presented in dots.
In addition to blood samples, we were able to collect 18 liver specimens from our BA cohort. Of these, blood and liver samples from 8 children shared identical variants (BA009, BA016, BA032, BA036, BA037, BA038, BA040 and BA041). Additionally, we did not detect any significant variants based on our rationales for variant classification (Table 3). In other words, this study did not detect any somatic variants from liver samples.
Table 3.
Identical variants detected from blood and liver samples.
| Proband | Sex (M/F) | Chr | Position | SNP ID | Ref | Alt | Gene | DNA change | A.A change | MOI | Genotype | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Proband | UM | UF | |||||||||||
| BA003 | F | Undetected | |||||||||||
| BA005 | M | Undetected | |||||||||||
| BA009 | M | X | 130,015,441 | rs201843717 | G | A | BCORL1 | NM_001184772:c.G2669 > A | p.Arg325Gln | X-linked | A | G/A | G |
| BA016 | M | X | 56,565,305 | C | G | UBQLN2 | NM_013444:c.C1432 > G | p.Pro478Ala | X-linked | G | C/G | C | |
| BA021 | F | Undetected | |||||||||||
| BA023 | M | Undetected | |||||||||||
| BA025 | F | Undetected | |||||||||||
| BA030 | M | Undetected | |||||||||||
| BA031 | M | Undetected | |||||||||||
| BA032 | M | 2 | 173,016,403 | rs773527960 | C | A | RAPGEF4 | NM_001282901:c.C1204 > A | p.Gln622Lys | AR | A/A | C/A | C/A |
| X | 129,590,191 | rs752439587 | T | A | OCRL | NM_001587:c.T2603 > A | p.Met876Lys | X-linked | A | T/A | T | ||
| BA035 | F | Undetected | |||||||||||
| BA036 | F | 16 | 75,243,074 | rs1327850193 | G | A | BCAR1 | NM_001170715:c.C83 > T | p.Ala10Val | De novo | G/A | G/G | G/G |
| BA037 | M | 9 | 20,948,857 | rs544335294 | C | A | FOCAD | NM_017794:c.C3805 > A | p.Pro1269Thr | AR | A/A | C/A | C/A |
| X | 70,341,839 | rs371383515 | A | C | KIF4A | NM_012310:c.A1174 > C | p.Asn392His | X-linked | C | A/C | A | ||
| BA038 | M | X | 47,448,875 | rs758443040 | G | A | ZNF41 | NM_001324139:c.C637 > T | p.Arg299Cys | X-linked | A | G/A | G |
| BA039 | F | Undetected | |||||||||||
| BA040 | M | X | 3,112,294 | rs1426850924 | C | T | ARSF | NM_001201538:c.C1511 > T | p.Pro504Leu | X-linked | T | C/T | C |
| X | 64,191,164 | rs764261510 | G | T | AMER1 | NM_152424:c.C2123 > A | p.Thr708Asn | X-linked | T | G/T | G | ||
| BA041 | M | 9 | 100,126,394 | rs148219510 | C | G | INVS | NM_014425:c.C118 > G | p.Leu40Val | AR | G/G | C/G | C/G |
| X | 129,557,351 | rs753369725 | G | C | OCRL | NM_000276:c.G265 > C | p.Asp89His | X-linked | C/C | G/C | G | ||
Chr chromosome, M male, F female, Ref reference, Alt alternative, A.A amino acid, MOI mode of inheritance, AR autosomal recessive, UM unaffected mother, UF unaffected father.
Effect of genetic predisposition
The detected variants showed extremely low MAFs against three employed databases: Kinh Vietnamese (KHV), GnomAD and 1000 Genome Project (Table 2). We noticed that the MAFs of the HACE1 and VPS13C variants were above 1% against the KHV database, while the rest were significantly below the thread hold of 1%. All variants with CADD Phred scaled scores were above 10 and mostly above 20, indicating either the 10% or 1% most deleterious substitutions, respectively. Among these variants, INVS:c.C208 > T was the most deleterious, with the highest scaled score of 37 (Table 2).
The Polyphen-2 and SIFT tools showed a consensus on the damaging impact of HACE1, PHKA1, XIAP, and AMER1 (c.A1075 > T), POF1B, MAOA, BCAR1, FOCAD, ARSF and OCRL variant, while the rest varied from tools (Table S1). We used I-Mutant to predict the stability of amino acid substitution for 28 identified variants via the change of free energy change values (DDG). The results show that except OCRL:c.T2603 > A(p.Met876Lys), which increased the stability of the mutant compared to that of the wild-type variants, all variants showed decreased stability (Table S2). The HOPE tool was used to predict the structural effect of missense variants, showing changes in residue size and hydrophobic and structural stability (Fig. S2). Changes in amino acid size and charge resulted in a loss of interaction and disturbance of protein function. Several variants, for example HACE1:c.G1660 > A(pAla651Thr) and PHKA1:c.G478 > A(p.Asp160Asn), whose wild-type residues are located in important domains. Thus, any substitution in these regions was predicted to lead to a functional disturbance. In contrast to I-mutant prediction, HOPE showed that an alternation of methionine by lysine residue in the variant OCRL:c.T2603 > A(p.Met876Lys) can disturb the hydrophobic interaction of the altered residue with other molecules on the surface of the protein (Fig. S2).
Analysis of biological function and human disease phenotype
Compute overlaps of 25 candidate genes to the human phenotype ontology from the Molecular Signatures Database, involving 4,494 gene sets (FDR q value < 0.05), indicated that the candidate genes felt into various human phenotype gene sets, ranging from gonosomal inheritance and X-linked recessive inheritance to involuntary movements (Table 4). We also computed our gene set to find the association of these genes with the reported phenotypes available from the HPO and Monarch Initiative (Table S3). However, we did not find any overlapping phenotypes from these databases. The reason might be a lack of genes/pathways associated with the BA phenotype in these available databases, which are often dominated by studies on Caucasians, where the prevalence of BA in this group is much lower than that in Asians. By applying the same strategy to identify the potential contribution of ciliary dysgenesis underlying the BA phenotype, we used a gene set containing 2016 genes of interest43. We found that some genes from our study, including BCOR, INVS and OCRL, were included in this gene set. This result suggested the novelty of BCOR, INVS and OCRL from our BA cohort.
Table 4.
Analysis of human phenotype ontology.
| Gene set name | # genes in gene set (K) | Description | # genes in overlap (k) | k/K | p value | FDR q-value | Gene overlap |
|---|---|---|---|---|---|---|---|
| HP_GONOSOMAL_INHERITANCE | 253 | Gonosomal inheritance | 12 | 0.0474 | 1.49E−20 | 6.70E−17 | ATRX, OCRL, THOC2, MAOA, KIF4A, BCORL1, XIAP, POF1B, PHKA1, BCOR, AMER1, UBQLN2 |
| HP_X_LINKED_RECESSIVE_INHERITANCE | 173 | X-linked recessive inheritance | 9 | 0.052 | 8.13 E−16 | 1.83 E−12 | ATRX, OCRL, THOC2, MAOA, KIF4A, BCORL1, XIAP, POF1B, PHKA1 |
| HP_SELF_INJURIOUS_BEHAVIOR | 108 | Self-injurious behaviour | 6 | 0.0556 | 5.66 E−11 | 8.47 E−08 | ATRX, OCRL, THOC2, MAOA, BCOR, ELP2 |
| HP_ABNORMAL_EMOTION_AFFECT_BEHAVIOR | 415 | Abnormal emotion/affect behaviour | 7 | 0.0169 | 4.97 E−09 | 4.97 E−06 | ATRX, OCRL, THOC2, MAOA, BCOR, ELP2, VPS13C |
| HP_NEUROLOGICAL_SPEECH_IMPAIRMENT | 1022 | Neurological speech impairment | 9 | 0.0088 | 6.24 E−09 | 4.97 E−06 | ATRX, OCRL, KIF4A, BCOR, AMER1, UBQLN2, ELP2, ZNF41HACE1 |
| HP_DELAYED_SPEECH_AND_LANGUAGE_DEVELOPMENT | 696 | Delayed speech and language development | 8 | 0.0115 | 6.63 E−09 | 4.97 E−06 | ATRX, THOC2, BCORL1, AMER1, ELP2, ZNF41, HACE1, CEP63 |
| HP_AUTISTIC_BEHAVIOR | 450 | Autistic behaviour | 7 | 0.0156 | 8.68 E−09 | 5.12 E−06 | ATRX, THOC2, MAOA, BCORL1, BCOR, VPS13C, ZNF41 |
| HP_ABNORMAL_AGGRESSIVE_IMPULSIVE_OR_VIOLENT_BEHAVIOR | 251 | Abnormal aggressive, impulsive or violent behaviour | 6 | 0.0239 | 9.12 E−09 | 5.12 E−06 | ATRX, OCRL, THOC2, MAOA, BCOR, ELP2 |
| HP_SHORT_STATURE | 1152 | Short stature | 9 | 0.0078 | 1.75 E−08 | 8.76 E−06 | ATRX, OCRL, THOC2, BCOR, AMER1, ELP2, HACE1, CEP63, INVS |
| HP_INVOLUNTARY_MOVEMENTS | 905 | Involuntary movements | 8 | 0.0088 | 5.05 E−08 | 2.19 E−05 | ATRX, OCRL, THOC2, MAOA, BCORL1, UBQLN2, ELP2, VPS13C |
Similar to a previous study43, we did not identify any variants in some genes that have been previously suggested to be associated with BA or BA-related diseases, such as PKD2 (polycystic kidney disease 2, polycystic kidney and hepatic disease 1), CFC1 (polysplenia), JAG1 (Alagille syndrome) and PKD1L1 (biliary atresia splenic malformation syndrome- BASM). We also did not find significant variants in the susceptibility loci of ADD3, XPNPEP1, GPC1, ARF6 and EFEMP1, as suggested by GWAS44.
Discussion
Similar to other previous studies, we attempted to reveal the genetic pattern of BA disorder by conducting trio-based exome sequencing for 40 families involving 41 children with BA. Going beyond this establishment in a genetic study for such a rare and complex disorder, we further tested our hypothesis of whether the detected variants occurred in somatic or germline cells by sequencing both blood and available liver specimens obtained from our BA cohort. Due to the complexity of BA, we applied a stringent bioinformatics pipeline and tight quality control to determine either the rarest variants or putative de novo events from our BA cohort, which would avoid a huge number of variants as often experienced from mass sequencing. Taking this straightforward principle enabled us to end up with a total of 28 variants in 25 respective genes. Identical variants detected from blood and liver samples allowed us to rule out the occurrence of somatic variants in the development of the disease as previously hypothesized45.
In agreement with previous studies, our results showed an intriguing genetic aspect of BA, which was highly heterogeneous. It is worth noting that along with other variants, this study found 3 genes whose variants occurred in unrelated probands, including AMER1, INVS and OCRL. While the aetiology of BA remained unclear and was unlikely to follow the Mendelian model, our results implicated their role in the disease's development. Overlapping findings of BCOR, INVS and OCRL in the Vietnamese BA cohort with a large comprehensive ciliopathy and biliary genes of interest in the previous study43 further supported the possibility of the causative role of these genes in BA. AMER1 (MIM#300647) encodes APC membrane recruitment protein 1, which acts as an inhibitor of the canonical Wnt/beta-catenin signalling pathway46 and controls hepatobiliary development during embryogenesis. In mature healthy liver cells, it is mostly inactive, and the abnormal Wnt/beta-catenin signalling pathway can promote the development of liver diseases47. AMER1 associates with osteopathia striata with cranial sclerosis48 and Wilms tumour development49–51. The gene is involved in the activation of the Wnt/beta-catenin signalling pathway, which drives hepatocarcinoma and cholangiocarcinoma52. In addition, analysis of the effect of genetic predispositions of AMER1 variants indicated that they were damaging because the alternated residues were located in highly conserved positions. The alternations might lead to destabilization of the local conformation and a loss of protein interaction (Table S1, S2, Fig. S2). Despite a lack of AMER1 to typical BA phenotypes, we inferred its indirect role in the development of BA as a result of activation of the Wnt/beta-catenin signalling pathway.
Our study highly suggested INVS as a BA candidate gene owing to INVS variant detection in 2 unrelated probands, their mode of inheritance and the effect of genetic predisposition. In particular, INVS: c.C208 > T (p.Arg396*) was de novo, and a loss-of-function variant with a CADD score of 37 and its allele frequency was absent from all employed databases. INVS encodes inversin protein, which plays a role in primary cilia function and is involved in the cell cycle. Intriguingly, inactivation of INVS in a mouse model shows a significant increase in bilirubin levels compared to that of the wild-type and pathogenic changes in ductal plate malformation in the intrahepatic biliary of the mutant mouse53. The association of INVS with BA had not been previously established due to an absence of INVS variants detected in BA patients54,55. However, INVS is associated with infantile nephronophthisis type 256–58. In our study, we detected an INVS heterozygous de novo variant and a homozygous variant from 2 BA unrelated patients (BA014 and BA041). To our knowledge, this novelty is first reported in BA patients, although future studies are needed to clearly explore the role of INVS in BA development. Similar to BCOR and INVS, OCRL encodes inositol polyphosphate 5-phosphatase, which might be involved in primary cilia assembly. OCRL has been widely reported to be linked to Lowe and Dent syndrome, where clinical manifestations often overlap with Zellweger spectrum disorders, characterized by low muscle tone, feeding difficulty, seizures and liver dysfunction59–61. Likewise, a lack of an association of OCRL and BA or liver diseases remains a gap for future investigation.
As a result of a rapidly declining cost of DNA sequencing, dozens of rare and previously undiagnosed genetic disorders are currently detectable. For the last 10 years, NGS technology has revolutionized our understanding of human genetics with a high level of accuracy, cost effectiveness and high throughput capability. NGS is steadily becoming a standard in routine diagnostic practices62. In BA studies, mitochondrial DNA has been found to associate with BA, suggesting the role of mitochondria in underling the pathogenic mechanism17. WES has revealed dozen candidate genes either encode ATP-binding cassette transporters (the ABC superfamily)18,19 or are involved in the Notch signalling pathway, such as JAG119,63 and NOTHC220. GWAS have highlighted a strong association between BA and some variants in the ADD3 gene located on 10q24.264. Another subsequent study on 171 BA patients and 1,630 controls of European descent found the strongest signal at rs7099604 in the ADD3 gene65. A significant association was found between variant rs17095355 on the XPNPEP1 gene and the disease66. Taken together, the aetiology of BA remains challenging due to the involvement of multiple genes and complex mechanisms. Being encouraged by the pioneers, we provided a concrete genetic aspect obtained from an exome trio-based study of a Vietnamese BA cohort. The findings add to our knowledge of the genetic heterogeneity and complexity of BA disorder.
Conclusion
The aetiology of BA remains challenging because there is a lack of conclusive evidence despite extensive research and medical practices for hundreds of years. However, the recent development of NGS technology and its application in studies of BA and liver diseases have gradually revealed the hidden genetic picture of BA aetiology, where dozens of BA-associated genes have been found. Our study identified 28 variants in 25 genes (all validated) from 41 children with BA. These variants were in the 10% most deleterious and were either rare or extremely rare in the population genome database. A combination of functional prediction and analysis of biological processes enabled us to suggest these candidate genes for the development of BA, particularly with those detected in unrelated BA individuals, including AMER1, INVS and OCRL. Identical variants detected from blood and liver wedge specimens from each BA individual suggested that somatic variants in the liver cells were unlikely to occur during morphogenesis. Taken together, we highlighted the genetic heterogeneity of BA and ruled out the Mendelian model. Future studies are needed to further explore the roles of these genes in the development of BA.
Supplementary Information
Author contributions
Conception of the study: K.T.T. and L.T.N. Patient recruitment and assessment: K.T.T.; H.K.N., L.T.N.; M.D.N.; H.T.N.; Q.A.T.; A.K.M. Genetic testing: K.T.T.; L.T.M.D.; H.K.N.; Bioinformatics analysis and data interpretation: V.S.L. and K.T.T. Manuscript preparation: all authors.
Funding
This study was funded by the Vinmec Healthcare System (Grant# ISC.18.13).
Data availability
Data are available from this manuscript and supplementary information.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Kien Trung Tran and Vinh Sy Le.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-021-01148-y.
References
- 1.Thomson J. On congenital obliteration of the bile-ducts. Edinb. Med. J. 1892;37:724–735. [PMC free article] [PubMed] [Google Scholar]
- 2.Kasai M. A new operation for “non-correctable” biliary atresia-Portoenterostomy. Shijitsu. 1959;13:733–739. [Google Scholar]
- 3.Davenport M, et al. The outcome of the older (≥ 100 days) infant with biliary atresia. J. Pediatr. Surg. 2004;39:575–581. doi: 10.1016/j.jpedsurg.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 4.Liu MB, et al. Biliary atresia in Vietnam: Management and the burden of disease. Surgery. 2017;161:533–537. doi: 10.1016/j.surg.2016.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Feldman AG, Mack CL. Biliary atresia: Clinical lessons learned. J. Pediatr. Gastroenterol. Nutr. 2015;61:167–175. doi: 10.1097/MPG.0000000000000755. [DOI] [PubMed] [Google Scholar]
- 6.Chardot C, et al. Improving outcomes of biliary atresia: French national series 1986–2009. J. Hepatol. 2013;58:1209–1217. doi: 10.1016/j.jhep.2013.01.040. [DOI] [PubMed] [Google Scholar]
- 7.Mezina A, Karpen SJ. Genetic contributors and modifiers of biliary atresia. Dig. Dis. 2015;33:408–414. doi: 10.1159/000371694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fallon SC, Chang S, Finegold MJ, Karpen SJ, Brandt ML. Discordant presentation of biliary atresia in premature monozygotic twins. J. Pediatr. Gastroenterol. Nutr. 2013;57:e22–23. doi: 10.1097/MPG.0b013e31826a1044. [DOI] [PubMed] [Google Scholar]
- 9.Ando K, et al. Sibling occurrence of biliary atresia and biliary dilatation. J. Pediatr. Surg. 1996;31:1302–1304. doi: 10.1016/S0022-3468(96)90259-6. [DOI] [PubMed] [Google Scholar]
- 10.Gunasekaran TS, Hassall EG, Steinbrecher UP, Yong SL. Recurrence of extrahepatic biliary atresia in two half sibs. Am. J. Med. Genet. 1992;43:592–594. doi: 10.1002/ajmg.1320430317. [DOI] [PubMed] [Google Scholar]
- 11.Lachaux A, et al. Familial extrahepatic biliary atresia. J. Pediatr. Gastroenterol. Nutr. 1988;7:280–283. doi: 10.1097/00005176-198803000-00020. [DOI] [PubMed] [Google Scholar]
- 12.Smith BM, Laberge JM, Schreiber R, Weber AM, Blanchard H. Familial biliary atresia in three siblings including twins. J. Pediatr. Surg. 1991;26:1331–1333. doi: 10.1016/0022-3468(91)90613-X. [DOI] [PubMed] [Google Scholar]
- 13.Muraji T, et al. Maternal microchimerism in underlying pathogenesis of biliary atresia: quantification and phenotypes of maternal cells in the liver. Pediatrics. 2008;121:517–521. doi: 10.1542/peds.2007-0568. [DOI] [PubMed] [Google Scholar]
- 14.Hayashida M, et al. The evidence of maternal microchimerism in biliary atresia using fluorescent in situ hybridization. J. Pediatr. Surg. 2007;42:2097–2101. doi: 10.1016/j.jpedsurg.2007.08.039. [DOI] [PubMed] [Google Scholar]
- 15.Davit-Spraul A, Baussan C, Hermeziu B, Bernard O, Jacquemin E. CFC1 gene involvement in biliary atresia with polysplenia syndrome. J. Pediatr. Gastroenterol. Nutr. 2008;46:111–112. doi: 10.1097/01.mpg.0000304465.60788.f4. [DOI] [PubMed] [Google Scholar]
- 16.Coffinier C, et al. Bile system morphogenesis defects and liver dysfunction upon targeted deletion of HNF1beta. Development (Cambridge, England) 2002;129:1829–1838. doi: 10.1242/dev.129.8.1829. [DOI] [PubMed] [Google Scholar]
- 17.Koh H, et al. Mitochondrial mutations in cholestatic liver disease with biliary atresia. Sci. Rep. 2018;8:905. doi: 10.1038/s41598-017-18958-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mezina A, et al. 845 whole exome sequencing identifies ABCB4 gene variants as modifiers of biliary atresia outcomes. Gastroenterology. 2014;146:S-928. doi: 10.1016/s0016-5085(14)63373-4. [DOI] [Google Scholar]
- 19.Sangkhathat S, Laochareonsuk W, Maneechay W, Kayasut K, Chiengkriwate P. Variants associated with infantile cholestatic syndromes detected in extrahepatic biliary atresia by whole exome studies: A 20-case series from Thailand. J. Pediatr. Genet. 2018;7:67–73. doi: 10.1055/s-0038-1632395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shaul E, et al. Novel mutations in NOTCH2 gene in infants with neonatal cholestasis. Pediatr. Rep. 2019;11:8206. doi: 10.4081/pr.2019.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bezerra JA, et al. Biliary atresia: Clinical and research challenges for the twenty-first century. Hepatology. 2018;68:1163–1173. doi: 10.1002/hep.29905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wildhaber BE. Biliary atresia: 50 years after the first kasai. ISRN Surg. 2012;2012:132089. doi: 10.5402/2012/132089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hsiao CH, et al. Universal screening for biliary atresia using an infant stool color card in Taiwan. Hepatology. 2008;47:1233–1240. doi: 10.1002/hep.22182. [DOI] [PubMed] [Google Scholar]
- 24.Fischler B, Haglund B, Hjern A. A population-based study on the incidence and possible pre- and perinatal etiologic risk factors of biliary atresia. J. Pediatr. 2002;141:217–222. doi: 10.1067/mpd.2002.126001. [DOI] [PubMed] [Google Scholar]
- 25.Liem NT, Son TN, Quynh TA, Hoa NP. Early outcomes of laparoscopic surgery for biliary atresia. J. Pediatr. Surg. 2010;45:1665–1667. doi: 10.1016/j.jpedsurg.2010.01.019. [DOI] [PubMed] [Google Scholar]
- 26.Luan PC, An PL, Phong NH, Ngoc NM. Characteristics of infants with cholestatic jaundice at gastrointestinal department, children’s hospital 2. Nghiên cứu Y học. 2014;18:402–407. [Google Scholar]
- 27.Sambrotta M, et al. Mutations in TJP2 cause progressive cholestatic liver disease. Nat. Genet. 2014;46:326–328. doi: 10.1038/ng.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tran KT, et al. Genetic landscape of autism spectrum disorder in Vietnamese children. Sci. Rep. 2020;10:5034. doi: 10.1038/s41598-020-61695-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Auton A, et al. A global reference for human genetic variation. Nature. 2015;526:68–74. doi: 10.1038/nature15393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997v2 [q-bio.GN] 1303 (2013).
- 31.McKenna A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H, et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. Vol. 1207 (2012).
- 34.Le VS, et al. A Vietnamese human genetic variation database. Hum. Mutat. 2019;40:1664–1675. doi: 10.1002/humu.23835. [DOI] [PubMed] [Google Scholar]
- 35.Rentzsch P, Witten D, Cooper GM, Shendure J, Kircher M. CADD: Predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 2019;47:D886–D894. doi: 10.1093/nar/gky1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaser R, Adusumalli S, Leng SN, Sikic M, Ng PC. SIFT missense predictions for genomes. Nat. Protoc. 2016;11:1–9. doi: 10.1038/nprot.2015.123. [DOI] [PubMed] [Google Scholar]
- 37.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2: Mutation prediction for the deep-sequencing age. Nat. Methods. 2014;11:361. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 39.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: Predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Venselaar H, Te Beek TA, Kuipers RK, Hekkelman ML, Vriend G. Protein structure analysis of mutations causing inheritable diseases. An e-Science approach with life scientist friendly interfaces. BMC Bioinform. 2010;11:548. doi: 10.1186/1471-2105-11-548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berauer JP, et al. Identification of polycystic kidney disease 1 like 1 gene variants in children with biliary atresia splenic malformation syndrome. Hepatology. 2019;70:899–910. doi: 10.1002/hep.30515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Girard M, Panasyuk G. Genetics in biliary atresia. Curr. Opin. Gastroenterol. 2019;35:73–81. doi: 10.1097/MOG.0000000000000509. [DOI] [PubMed] [Google Scholar]
- 45.Fabre A, Roman C, Roquelaure B. Somatic mutation, a cause of biliary atresia: A hypothesis. Med. Hypotheses. 2017;102:91–93. doi: 10.1016/j.mehy.2017.03.015. [DOI] [PubMed] [Google Scholar]
- 46.Tanneberger K, et al. Structural and functional characterization of the Wnt inhibitor APC membrane recruitment 1 (Amer1) J. Biol. Chem. 2011;286:19204–19214. doi: 10.1074/jbc.M111.224881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Perugorria MJ, et al. Wnt-beta-catenin signalling in liver development, health and disease. Nat. Rev. Gastroenterol. Hepatol. 2019;16:121–136. doi: 10.1038/s41575-018-0075-9. [DOI] [PubMed] [Google Scholar]
- 48.Holman SK, et al. Osteopathia striata congenita with cranial sclerosis and intellectual disability due to contiguous gene deletions involving the WTX locus. Clin. Genet. 2013;83:251–256. doi: 10.1111/j.1399-0004.2012.01905.x. [DOI] [PubMed] [Google Scholar]
- 49.Rivera MN, et al. An X chromosome gene, WTX, is commonly inactivated in wilms tumor. Science. 2007;315:642. doi: 10.1126/science.1137509. [DOI] [PubMed] [Google Scholar]
- 50.Ruteshouser EC, Robinson SM, Huff V. Wilms tumor genetics: Mutations in WT1, WTX, and CTNNB1 account for only about one-third of tumors. Genes Chromosomes Cancer. 2008;47:461–470. doi: 10.1002/gcc.20553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Akhavanfard S, et al. Inactivation of the tumor suppressor WTX in a subset of pediatric tumors. Genes Chromosomes Cancer. 2014;53:67–77. doi: 10.1002/gcc.22118. [DOI] [PubMed] [Google Scholar]
- 52.Carotenuto P, et al. Wnt signalling modulates transcribed-ultraconserved regions in hepatobiliary cancers. Gut. 2017;66:1268–1277. doi: 10.1136/gutjnl-2016-312278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shimadera S, et al. The inv mouse as an experimental model of biliary atresia. J. Pediatr. Surg. 2007;42:1555–1560. doi: 10.1016/j.jpedsurg.2007.04.018. [DOI] [PubMed] [Google Scholar]
- 54.Rajagopalan R, et al. Exome sequencing in individuals with isolated biliary atresia. Sci. Rep. 2020;10:2709. doi: 10.1038/s41598-020-59379-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schon P, et al. Identification, genomic organization, chromosomal mapping and mutation analysis of the human INV gene, the ortholog of a murine gene implicated in left-right axis development and biliary atresia. Hum. Genet. 2002;110:157–165. doi: 10.1007/s00439-001-0655-5. [DOI] [PubMed] [Google Scholar]
- 56.Halbritter J, et al. Identification of 99 novel mutations in a worldwide cohort of 1056 patients with a nephronophthisis-related ciliopathy. Hum. Genet. 2013;132:865–884. doi: 10.1007/s00439-013-1297-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kang HG, et al. Targeted exome sequencing resolves allelic and the genetic heterogeneity in the genetic diagnosis of nephronophthisis-related ciliopathy. Exp. Mol. Med. 2016;48:e251. doi: 10.1038/emm.2016.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schueler M, et al. Large-scale targeted sequencing comparison highlights extreme genetic heterogeneity in nephronophthisis-related ciliopathies. J. Med. Genet. 2016;53:208–214. doi: 10.1136/jmedgenet-2015-103304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Coon BG, et al. The Lowe syndrome protein OCRL1 is involved in primary cilia assembly. Hum. Mol. Genet. 2012;21:1835–1847. doi: 10.1093/hmg/ddr615. [DOI] [PubMed] [Google Scholar]
- 60.Luo N, et al. OCRL localizes to the primary cilium: A new role for cilia in Lowe syndrome. Hum. Mol. Genet. 2012;21:3333–3344. doi: 10.1093/hmg/dds163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang X, Jefferson AB, Auethavekiat V, Majerus PW. The protein deficient in Lowe syndrome is a phosphatidylinositol-4,5-bisphosphate 5-phosphatase. Proc. Natl. Acad. Sci. U S A. 1995;92:4853–4856. doi: 10.1073/pnas.92.11.4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nicastro E, D'Antiga L. Next generation sequencing in pediatric hepatology and liver transplantation. Liver Transpl. 2018;24:282–293. doi: 10.1002/lt.24964. [DOI] [PubMed] [Google Scholar]
- 63.Fischetto R, et al. Alagille syndrome: A novel mutation in JAG1 gene. Front. Pediatr. 2019;7:199. doi: 10.3389/fped.2019.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng G, et al. Common genetic variants regulating ADD3 gene expression alter biliary atresia risk. J. Hepatol. 2013;59:1285–1291. doi: 10.1016/j.jhep.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 65.Tsai EA, et al. Replication of a GWAS signal in a Caucasian population implicates ADD3 in susceptibility to biliary atresia. Hum. Genet. 2014;133:235–243. doi: 10.1007/s00439-013-1368-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garcia-Barcelo MM, et al. Genome-wide association study identifies a susceptibility locus for biliary atresia on 10q24.2. Hum. Mol. Genet. 2010;19:2917–2925. doi: 10.1093/hmg/ddq196. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data are available from this manuscript and supplementary information.