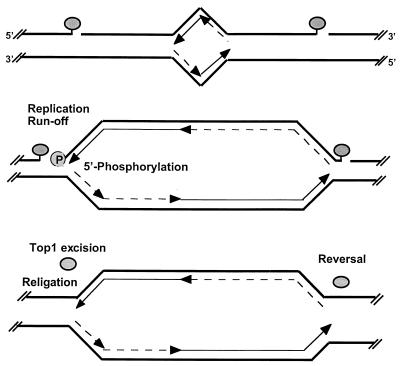
FIG. 9.
Proposed interactions of DNA replication forks with CPT-stabilized top1 cleavage complexes and hypothetical repair pathways. Two covalent top1 cleavage complexes (shaded ovals) are shown, one on each side of a growing replication bubble (top). Parental DNA strands are represented as thick lines. Leading-strand synthesis is shown as thin arrows, and Okazaki fragments are shown as broken-line arrows. The differential effect of replication fork collision into top1 cleavage complexes on the leading and lagging strands is shown in the middle panel. Our results suggest that replication-mediated DNA double-strand breaks are formed by replication fork runoff on the leading strand with phosphorylation (P) of the 5′ end of the DNA template strand. By contrast, replication-mediated DNA double-strand breaks are not detectable on the lagging strand, which suggests that the replication fork is arrested upstream from the top1 cleavage complex without bypass, that the replication complex forces the dissociation of the top1 cleavage complex, or that Okasaki fragment synthesis bypasses the top1-mediated single-strand break interruption (see Discussion). In any case, no replication runoff would occur on the replicating lagging strand. (Bottom) Hypothetical excision repair of top1 cleavage complexes on the leading strand (see text for details and references). On the lagging strand, top1 might religate the DNA template strand directly upon drug removal. Replication-mediated DNA double-strand breaks are potential targets for homologous and illegitimate recombination.