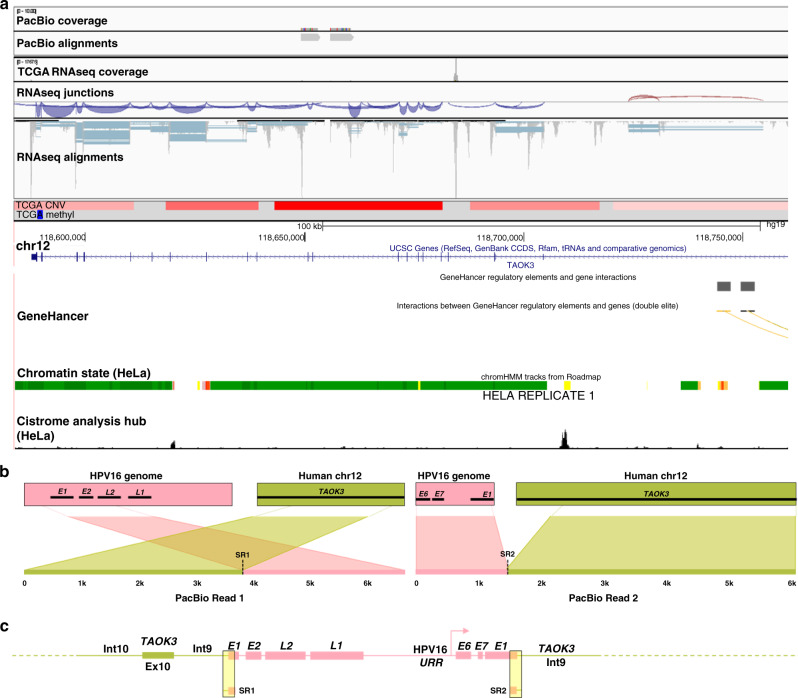
Fig. 5. Annotation of HPV integration affecting TAOK3 expression in TCGA-C5-A2LX.
PacBio long-read sequencing, TCGA, and UCSC Genome Browser (http://genome.ucsc.edu) data were used to annotate the HPV integration site within the candidate IDG, TAOK3. TCGA-C5-A2LX long-read data (PacBio) and TCGA sequencing (RNAseq), CNV (blue = loss; red = gain), and methylation data (blue = hypomethylation; red = hypermethylation) covering integration event is depicted in panel (a). The Ribbon programme was used to generate a schematic of two PacBio reads covering the area of integration. Thick bars across the top (b) represent the HPV and human reference genomes, which are connected by dashed lines to two unique PacBio reads covering the integration to show how they are specifically mapped to each genome. Data from all PacBio long reads covering the integration event were used to hand annotate the integration event (c). Breakpoints identified from TCGA short-read sequencing (SR) are highlighted in the yellow boxes. PacBio sequencing successfully captured the entirety of the HPV insertion, which comprised almost two full copies of the HPV16 genome (pink) flanked on both sides by intron 9 of TAOK3 (olive).