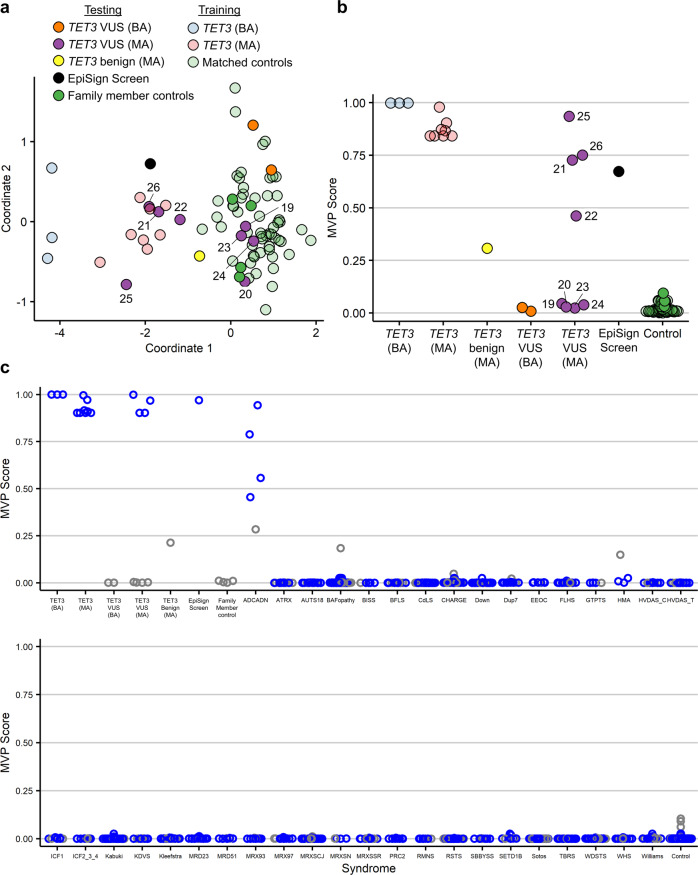
Fig. 3. Classification of samples using the TET3 episignature.
a Multidimensional scaling (MDS) plot of the TET3 signature discovery, validation, and testing samples, including one previously unknown sample identified from the EpiSign database, using the episignature from the second round of signature discovery. The discovery and validation pathogenic samples (n = 3 TET3 (BA), light blue; n = 8 TET3 (MA), light red) along with matched controls (n = 55, light green) were used to identify a TET3 episignature, which was then used to classify the remaining samples, including the VUS’s of the testing cohort (n = 2 TET3 VUS (BA), orange; n = 8 TET3 VUS (MA), purple; n = 1 TET3 benign (MA), yellow; n = 1 EpiSign screen, black; n = 4 family member controls, green). The 66 samples used for signature identification are shown as partially transparent circles, and the remaining samples are opaque. See Table 1 for descriptions of samples. TET3 VUS (MA) samples are numbered according to Table 1. b Methylation variant pathogenicity (MVP) plot of the same samples. c The TET3 episignature from the final round of signature discovery applied to samples from 46 other neurodevelopmental conditions, which exhibit 38 different DNA methylation episignatures in our EpiSign database (some syndromes share signatures). For each syndrome and for control samples, 75% of samples were used to train the classifier (blue) and 25% were used for testing (gray). VUS, variants of uncertain significance; TET3 VUS (BA), samples with bi-allelic TET3 VUS’s; TET3 VUS (MA), samples with mono-allelic TET3 VUS’s; TET3 benign (MA), the benign variant that did not reduce catalytic activity in vitro8; EpiSign screen, an unknown sample identified by screening the Episign database; family member controls, family members of affected individuals lacking TET3 variants; TET3 (BA), samples with bi-allelic pathogenic TET3 variants; TET3 (MA), samples with mono-allelic pathogenic TET3 variants; matched controls, age-matched and sex-matched controls. Syndrome abbreviations: ADCADN, Autosomal dominant cerebellar ataxia, deafness, and narcolepsy; ATRX, Alpha-thalassemia mental retardation syndrome; AUTS18, Autism, susceptibility to, 18; BAFopathy, Coffin-Siris 1–4,8 (CSS1–4,8) & Nicolaides-Baraitser (NCBRS) syndromes; BISS, Blepharophimosis Intellectual disability SMARCA2 Syndrome; BFLS, Börjeson–Forssman–Lehmann syndrome; CdLS, Cornelia de Lange syndrome; CHARGE, CHARGE syndrome; Down, Down syndrome; Dup7, Williams-Beuren region duplication syndrome (Chr7q11.23 duplication syndrome); EEOC, epileptic encephalopathy, childhood-onset; FLHS, Floating-Harbor syndrome; GTPTS, Genitopatellar syndrome; HMA, Hunter-McAlpine syndrome; HVDAS_C, Helsmoortel-van der Aa syndrome (ADNP syndrome [Central region methylation signature]); HVDAS_T, Helsmoortel-van der Aa syndrome (ADNP syndrome [Terminal regions methylation signature]); ICF1, Immunodeficiency-centromeric instability-facial anomalies syndrome Type 1; ICF2_3_4, Immunodeficiency-centromeric instability-facial anomalies syndrome Types 2, 3, 4; Kabuki, Kabuki syndrome 1 and 2; KDVS, Koolen de Vreis syndrome; Kleefstra, Kleefstra syndrome; MRD23, mental retardation autosomal dominant 23; MRD51, mental retardation autosomal dominant 51; MRX93, mental retardation X-linked 93; MRX97, mental retardation X-linked 97; MRXSCJ, mental retardation X-linked, syndromic, Claes-Jensen type; MRXSN, mental retardation X-linked syndromic Nascimento-type; MRXSSR, mental retardation X-linked Snyder-Robinson type; PRC2, PRC2 complex (Weaver syndrome and Cohen-Gibson syndrome); RMNS, Rahman syndrome; RSTS, Rubinstein-Taybi syndrome; SBBYSS, Ohdo syndrome, SBBYSS variant; SETD1B, SETD1B-related syndrome; Sotos, Sotos syndrome; TBRS, Tatton-Brown-Rahman syndrome; WDSTS, Wiedemann-Steiner syndrome; WHS, Wolf-Hirschhorn syndrome; Williams, Williams-Beuren syndrome (Chr7q11.23 deletion syndrome).