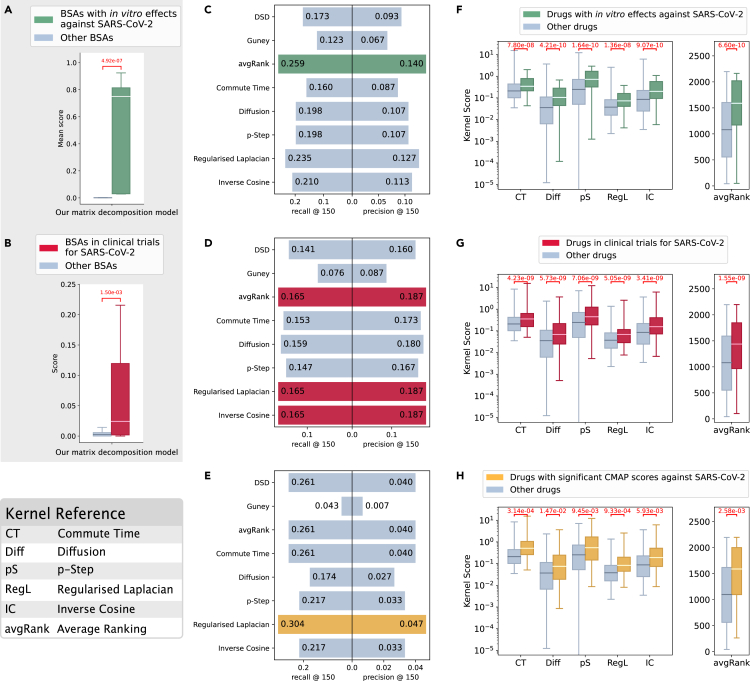
Figure 5.
Analysis of the predictions for COVID-19
We used three different sources of evidence: in vitro (A, C, and F), clinical trials (B, D, and G), and CMAP (E and H). We compared scores for drugs with evidence of efficacy against SARS-CoV2 versus scores for the remaining drugs. Our matrix factorization model (A and B) and kernel-based methods (F, G, and H) provide scores that are significantly different between the two groups of drugs in every case (Wilcoxon-Mann-Whitney p < 0.05). We formulated a binary classification problem to discriminate between drugs with evidence of efficacy against SARS-CoV2 and the remaining drugs. (C, D, and F) Comparison of precision and recall at top 150 for our kernel-based methods (commute time, diffusion, p-step, regularized Laplacian, and inverse cosine kernels, and avgRank), DSD, and Guney’s distance. The highest values are colored.