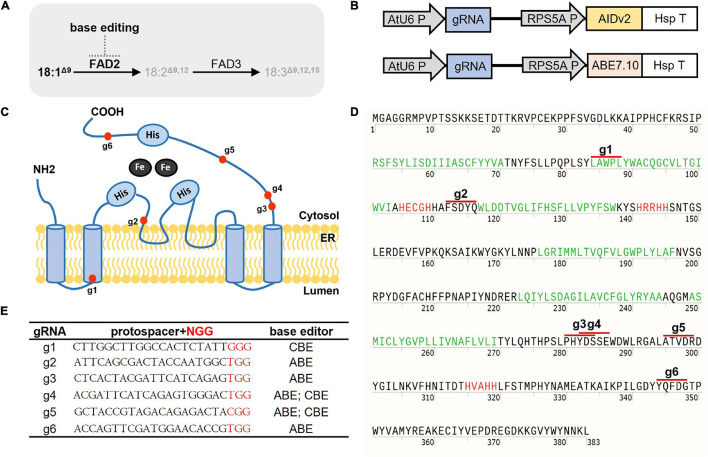
FIGURE 1.
“Semi-random” base editing strategy for selected FAD2 coding regions. (A) The function of FAD2 in desaturation of fatty acids and potential BE action in Arabidopsis seeds. FAD2 desaturates 18:1Δ9 to 18:2Δ9,12. FAD2 is subjected to base editing to alter its function. (B) Core structures of the CRISPR part in T-DNA from binary vectors harboring CBE and ABE in this study. gRNA expression is controlled by the U6 promoter, and AIDv2-dependent CBE and ABE7.10 are under the control of RPS5A promoters. (C) A schematic diagram of the FAD2 protein structure adapted from Zhang et al. (2012) ER-located FAD2 has six transmembrane domains and three histidine box motifs (His). Red dots denote BE target regions. (D) The FAD2 amino-acid sequence. Arabidopsis FAD2 is composed of 383 amino acids. Green-colored sequences indicate transmembrane domains, red-colored sequences indicate His motifs, and red lines indicate the potential amino acids that might be affected by BE targeting via corresponding gRNAs. (E) A list of selected gRNAs used for CBE and/or ABE with a protospacer sequence for FAD2 editing in this study.