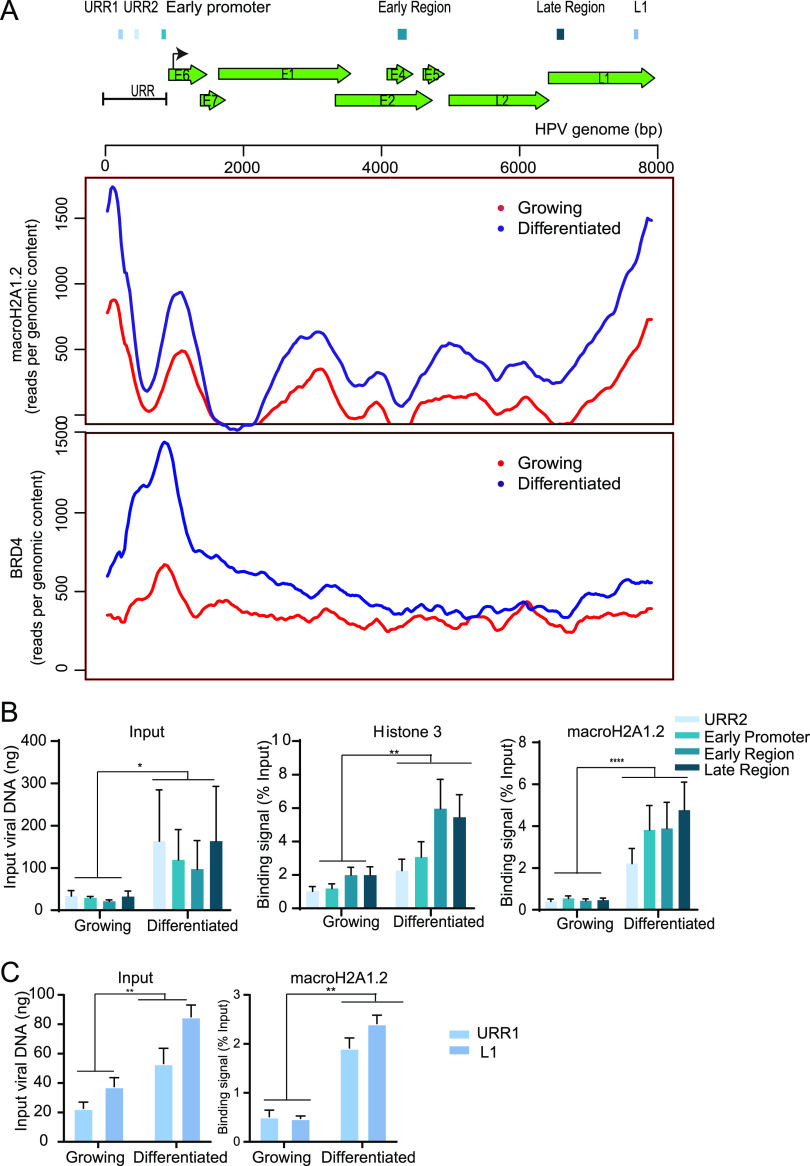
FIG 3.
macroH2A1 and Brd4 binding to the HPV31 genome is increased in differentiated conditions. (A) Schematic of a linearized HPV31 genome. URR, upstream regulatory region. ChIPseq was performed with macroH2A1.2 and Brd4 antibodies. Alignment of ChIPseq reads to the HPV31 reference genome in samples from growing and differentiated 9E cells is shown. ChIPseq reads were aligned and analyzed. Data were averaged from two biological replicates. (B) ChIP signals for histone H3 and macroH2A1.2 were expressed as a percentage of immunoprecipitated viral DNA relative to the total amount of input chromatin. Background signal (measured by immunoprecipitating viral DNA with IgG antibody) was subtracted from the corresponding ChIP signals. Binding levels were averaged from three independent experiments. Error bars represent ± standard error of the mean (SEM), and statistical significance was calculated using a paired Student’s t test. (C) ChIP signals were expressed as a percentage of immunoprecipitated viral DNA relative to the total amount of input chromatin. Background signal (measured by immunoprecipitating viral DNA with IgG antibody) was subtracted from the corresponding ChIP signals. URR1 and L1 regions were selected from ChIPseq peaks, and ChIP was carried out independent of the ChIP experiment shown in panel B. Binding levels were averaged from two independent experiments. Error bars represent ± standard deviation (SD), and statistical significance was calculated using a paired Student’s t test. *, P < 0.05; **, P < 0.005; ***, P < 0.005.