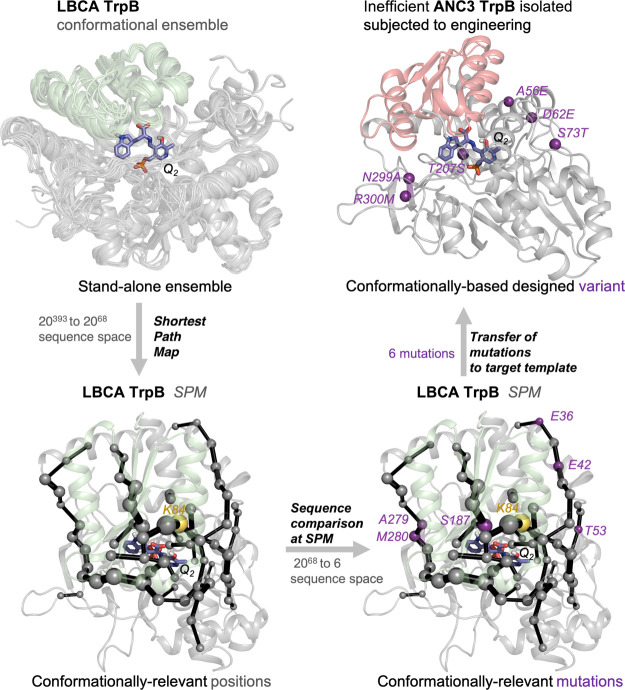
Figure 3.
SPM-based computational workflow for SPM6 TrpB enzyme variant generation. By analyzing the conformational ensemble of the stand-alone LBCA TrpB with high catalytic activity (upper left ensemble) through the SPM, we identified positions (gray spheres, lower left structure) within allosteric pathways (black edges) in the enzyme that contribute most to the LBCA TrpB conformational dynamics in the Q2 intermediate. Thereby the size of each edge and node corresponds to the relevance for conformational dynamics. The catalytic residue K84 is highlighted in yellow. Excluding residues that do not participate in an allosteric pathway reduces the sequence space from 20393 to 2068 possible activity enhancing substitutions. Sequence comparison at the SPM positions between stand-alone LBCA TrpB and inefficient ANC3 TrpB reduces the sequence space to six mutations with respect to LBCA TrpB (lower right structure, purple residues), that were introduced into ANC3 TrpB (upper right structure, purple residues) and tested in vitro. Numbering of the residues is according to LBCA TrpB in the lower right panel and according to ANC3 TrpB in the upper right panel. Note that an insertion of 20 amino acids in ANC3 TrpB relative to LBCA TrpB leads to the shift in the residue numbering.