Figure 2.
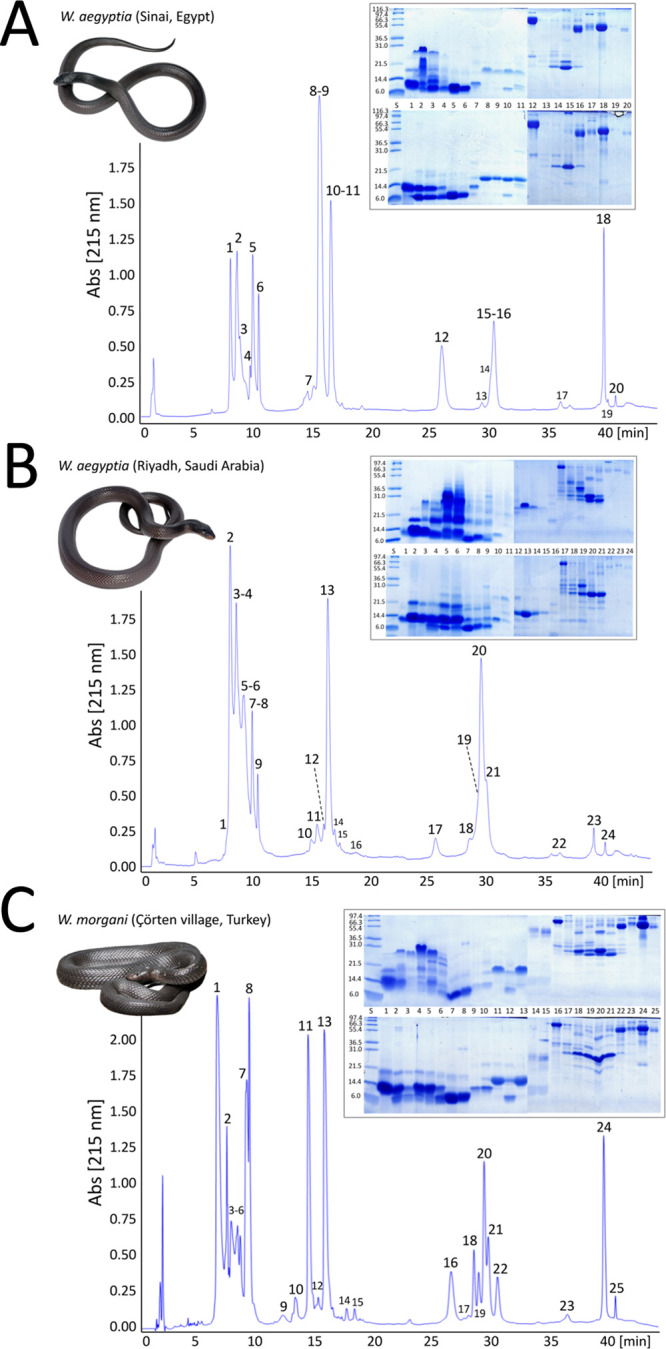
Bottom-up venomics analysis of the toxin arsenal of desert black cobras, W. aegyptia and W. morgani. Panels (A–C) display, respectively, reverse-phase chromatographic separations of the venom proteins of two W. aegyptia specimens (Sinai Peninsula, Egypt, and Riyadh, Saudi Arabia) and a venom sample from a W. morgani specimen original from Çörten village (Turkey). For venomics analyses, chromatographic fractions were collected manually and analyzed by SDS-PAGE (inset) under nonreduced (upper panels) and reduced (lower panels) conditions. Protein bands were excised, in-gel digested with trypsin, and the resulting proteolytic peptides were fragmented through LC-nESI-MS/MS. Parent proteins were identified by database searching (against the last update of the NCBI nonredundant database, including the W. aegyptia venom gland transcriptomic data deposited with the SRA and TSA databases, Supporting Information Table S1) and de novo sequencing followed by BLAST analysis (Supporting Information Tables S2–S4). Picture of W. aegyptia specimens displayed in panels (A) and (B) were taken by Salvador Carranza. Picture of W. morgani, Bayram Göçmen.
