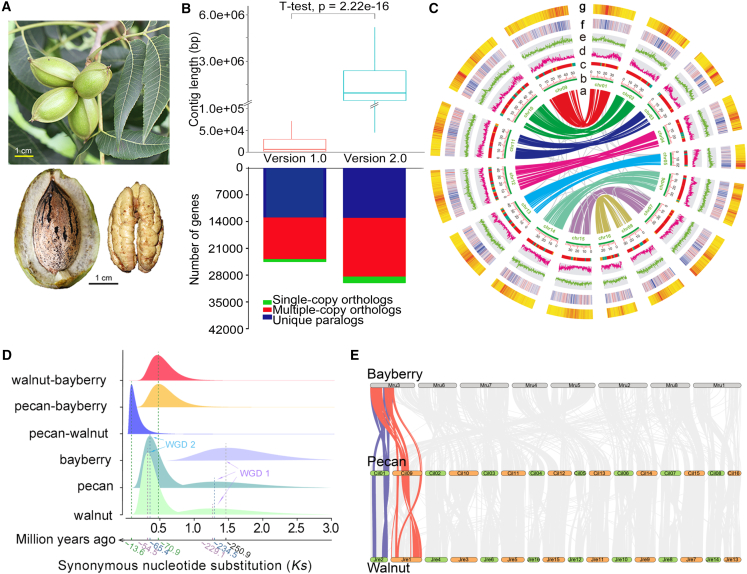
Figure 1.
Genome features of the chromosome-scale assembly and evolution of the pecan genome.
(A) Morphology of fruit, nut, and fresh kernel of Pawnee. Scale bar corresponds to 1 cm.
(B) Comparison of contig lengths and clustered gene sets between versions 1.0 and 2.0.
(C) Landscape of the chromosome-scale pecan genome assembly. (a) Synteny of gene pairs from the recent WGD (WGD 2); (b) chromosomes; (c) contigs of version 1.0 on chromosomes of version 2.0; red, green, and yellow indicate contigs >2, 1–2, and <1 Mb in length in the version 1.0 assembly; (d) protein-coding genes; (e) LTR density distribution; (f–g) distribution of Copia and Gypsy elements.
(D) WGD and divergence within and among species of pecan, walnut, and bayberry.
(E) Syntenic distribution of pecan compared with walnut and bayberry.