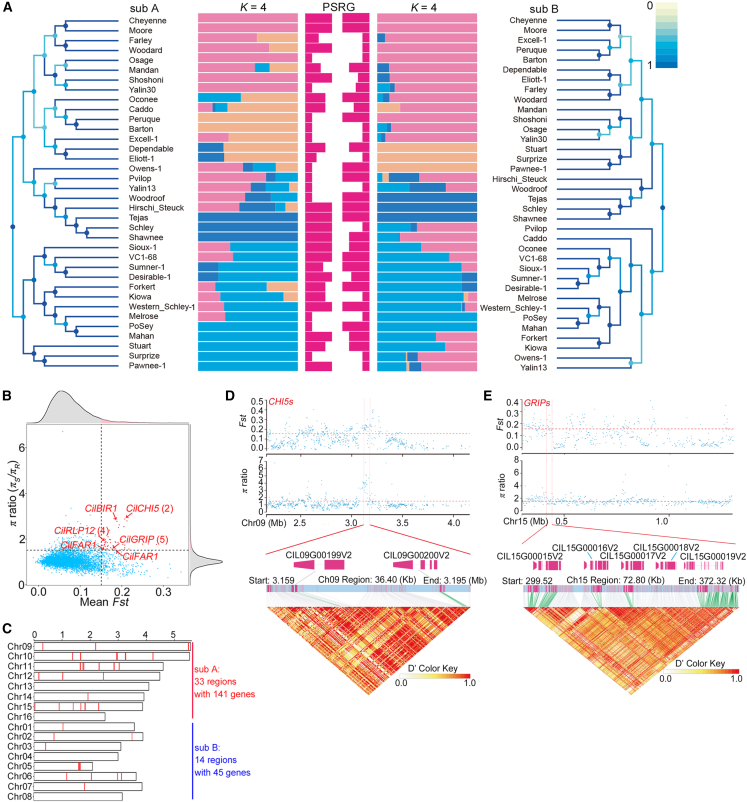
Figure 3.
Population genomics and identification of pecan scab-associated candidate genomic regions and genes under selection.
(A) NJ trees and population structures of subgenomes and scab-resistance grades of each accession. A score of 1 on the color scale bar indicates that the tree structure of the node is identical to the tree structure of its best corresponding node. K = 4, the best substructure. PSRG, pecan scab-resistance grade. The length of the red bars in the middle indicates the disease-resistance grade.
(B) Plots of the highest 5% π and Fst values in pecan scab-related accessions. Arrowheads indicate the loci of key candidate genes associated with pecan scab resistance.
(C) Locations of the selected regions of pecan scab resistance on chromosomes.
(D)Fst and π ratio (upper, coordinate diagrams), gene details (middle, color bars), and LD heatmap of the candidate region containing two putative chitinase-encoding genes.
(E)Fst and π ratio (upper, coordinate diagrams), gene details (middle, color bars), and LD heatmap of the candidate region containing five putative GRIP-encoding genes. The pairwise LD between the SNPs is indicated as D′ values, where red indicates a value of 1 and yellow indicates 0. Rose-red indicates the CDS regions of genes, and green indicates polymorphic SNP sites in the promoter and CDS regions.