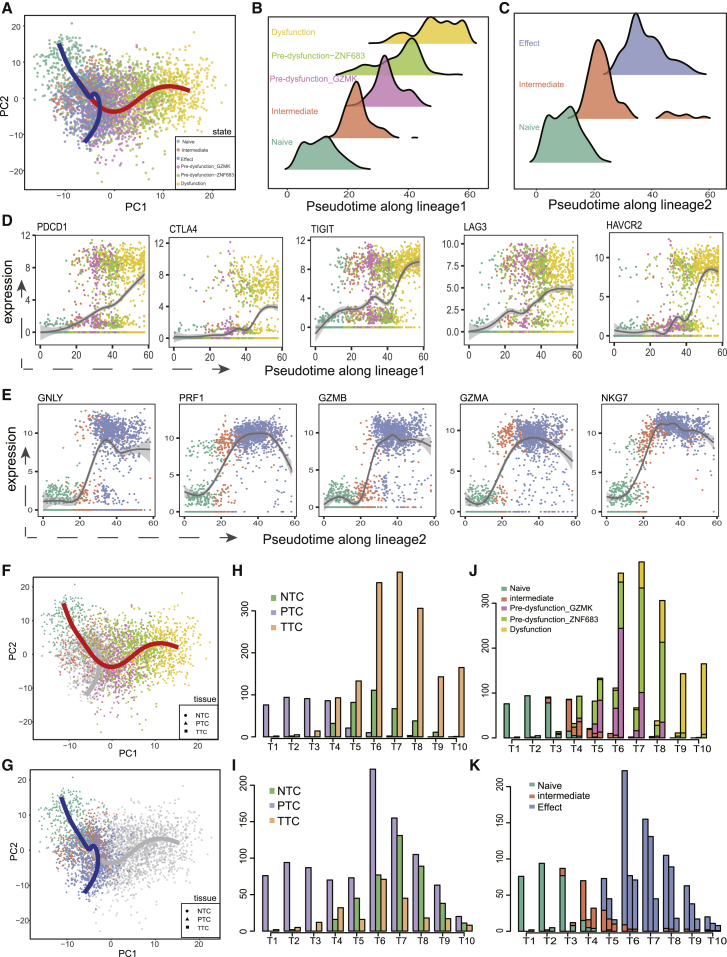
Figure 1.
scRNA-seq revealed a bifurcated trajectory of CD8+ T cells
(A) Pseudo-time analysis of CD8+ T cells using Slingshot algorithm, resulting in two lineages (colored lines) in the trajectory. Cells on the trajectory are labeled with cell states, revealing a dysfunction lineage (red line) versus effect lineage (blue line). (B and C) Pseudo-time density distribution of each state on the dysfunction lineage (B) and on the effect lineage (C). (D) Expression levels of co-inhibitory receptors along pseudo-time of the dysfunction lineage. (E) Expression levels of effector molecules along pseudo-time of the effect lineage. (F and G) Tissues of origin for cells on the dysfunction lineage (F) and on the effect lineage (G). Points shaped by ellipse, triangle, and rectangle denote PTC, NTC, and TTC, which were short for CD8+ T cells from peripheral blood, adjacent normal tissues, and tumor tissues, respectively. (H and J) Pseudo-time distribution (H) and state composition (J) of cells from different origins of tissues on the dysfunction lineage. The lineage is divided evenly into 10 bins according to pseudo-time. The height of bars represents the number of cells from various tissues in each bin. (I and K) Pseudo-time distribution (I) and state composition (K) of cells from different origins of tissues on the effect lineage.