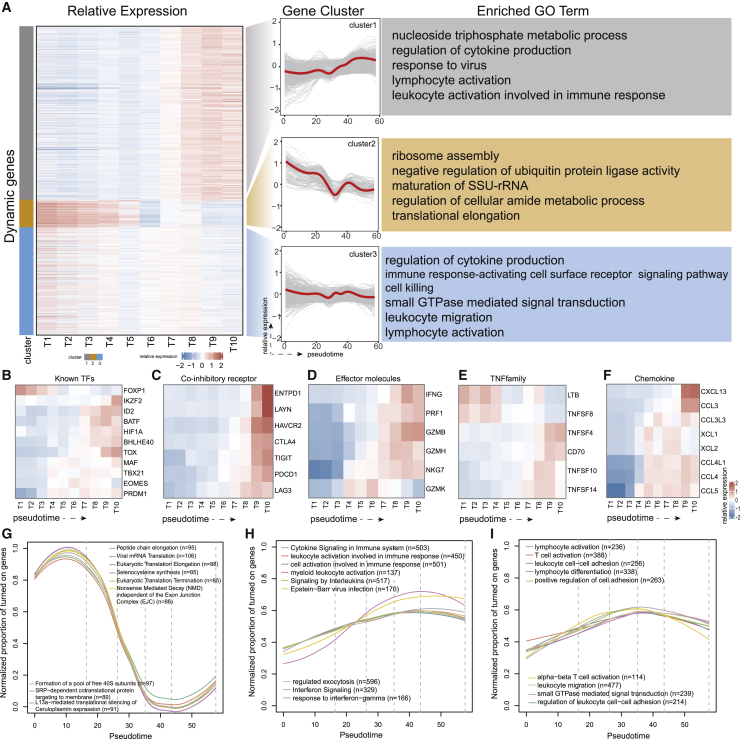
Figure 2.
Dynamic gene analysis on the dysfunction lineage
(A) (Left) Heatmap showing average expression of dynamic genes in 10 bins of cells evenly divided according to pseudo-time. All of the dynamic genes were grouped into three clusters. (Middle) Expression trends along the dysfunction lineage for all genes in each cluster (gray lines) and for the cluster average (red lines). (Right) Representative enriched Gene Ontology (GO) terms for each cluster. (B–F) Dynamic changes in the expression of representative genes contained in dynamic genes, including known TFs (B), co-inhibitory receptors (C), effector molecules (D) tumor necrosis factor family (TNF family) (E), and chemokines (F). (G–I) Dynamic changes in the activities of function entities for cluster 2 (G), cluster 1 (H), and cluster 3 (I). The top 10 enriched terms for each cluster were displayed.