Figure 3.
Constructing and validating dynamic networks along the dysfunction lineage
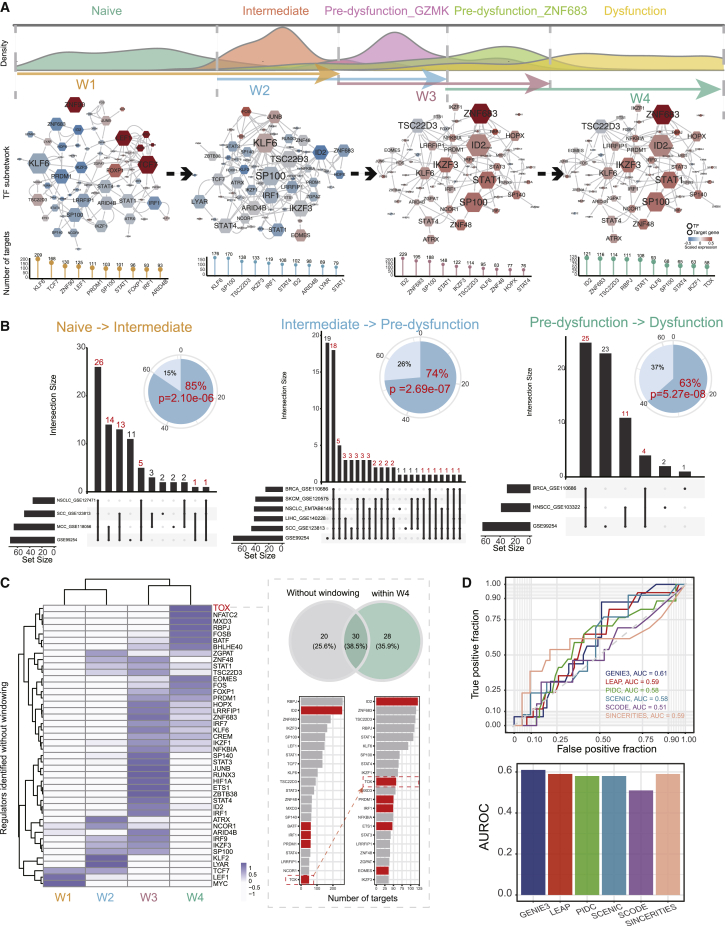
(A) (Upper) Pseudo-time densities for cells in different states (naive, intermediate, pre-dysfunction_GZMK, pre-dysfunction_ZNF683, dysfunction). The cells in two consecutive adjacent states were windowed to construct the gene regulatory network (GRN), which reflected the transition between the two states. (Middle) The sub-networks of TFs in each window. For each network, nodes are colored by average expression in the corresponding window. The sizes of nodes were scaled by their numbers of target genes. (Lower) The lollipop plot displays the number of target genes for the top 10 regulators. (B) Overlap of regulators identified in different datasets for each state transition. The pie plot displays the proportions of regulators identified only in the core dataset (light blue) and identified in at least one independent dataset (blue). The significance was evaluated by a hypergeometric test. (C) The heatmap displays the scaled Jaccard index estimating overlap of targets for each TF in each window and that in the network without windowing. The Venn diagram displays the overlap of target genes of the TOX in network W4 and network without windowing. The number of target genes for top regulators in the two networks were listed, and curated TFs related to T cell dysfunction were colored red. (D) Receiver operating characteristic (ROC) curve and area under the ROC (AUC) depicting the performance on prediction of dysfunction-related regulatory factors for six GRN construction algorithms.