Figure 3.
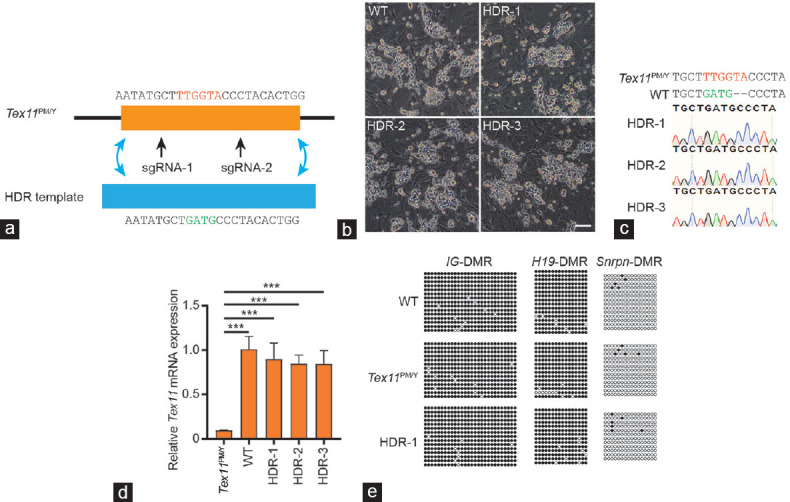
Correction of Tex11 mutation via CRISPR-Cas9-mediated gene editing in SSCs. (a) Schematic for Tex11 gene correction via HDR induced by CRISPR-Cas9 system and HDR template with WT sequence of Tex11. The mutant nucleotides were marked in red, while the WT nucleotides were in green. (b) Morphology of the three established SSC lines with correct Tex11 sequence (termed HDR-1, 2, and 3). Scale bar = 100 μm. (c) DNA sequencing analysis of SSCs from HDR-1/2/3. Note that the sequences of PCR products amplified from the Tex11 gene show that HDR-1/2/3 carry corrected Tex11 gene. (d) Transcriptional analysis of Tex11 in the corrected SSCs lines. The expression values were normalized to that of WT. The average values of three separate experiments are plotted; error bar: s.d. Significance was determined by two-tailed, unpaired Student’s t-test; ***P < 0.001. (e) DNA methylation analysis of the DMRs of H19, IG, and Snrpn in Tex11PM/Y and HDR-1 SSCs. Open, filled, and gray circles represent unmethylated, methylated, and uncertain CpG sites, respectively. WT: wild-type; Tex11: testis expressed 11; SSCs: spermatogonial stem cells; s.d.: standard deviation; sgRNA: single-guide RNA; CRISPR-Cas9: clustered regularly interspaced short palindromic repeats-CRISPR-associated endonuclease 9; DMR: differentially methylated regions; H19: H19 imprinted maternally expressed transcript; IG: intergenic differentially methylated region; Snrpn: small nuclear ribonucleoprotein polypeptide N; CpG: cytosine-guanine; HDR: homology-directed repair.
