Figure 4.
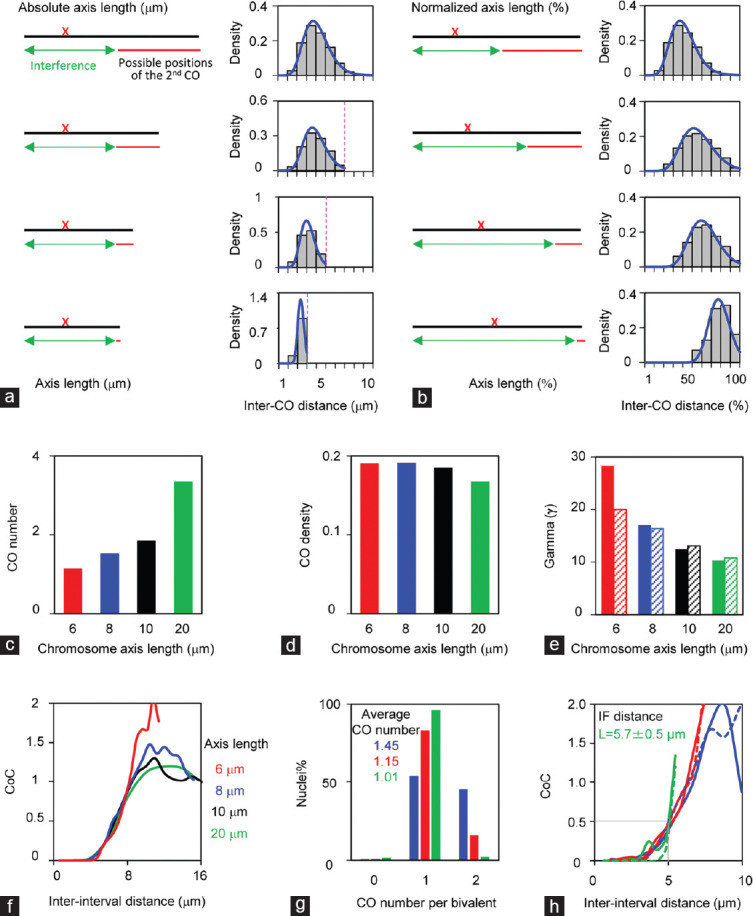
Chromosome axis length regulates crossover patterns. (a–f) Simulated data from four artificial chromosomes with different axis lengths (20 μm, 10 μm, 8 μm, and 6 μm) to illustrate the effects of chromosome lengths on crossover patterns. The same set of parameters used for all chromosomes: CO designation driving force, Smax= 4.1; clamped left and right ends, cL = cR = 1; the evenness level of precursors along chromosomes, E = 0.6; precursor distribution among chromosomes, B = 0.6; interference distance, L = 5 μm; precursor number N is adjusted according to axis length (N= 20, 10, 8, and 6, separately). (a and b) Illustration of how short chromosomes alter crossover number and distribution. Chromosome axis lengths are (a) shown as microns or (b) normalized to “1.” (c–e) Short chromosomes have (c) fewer crossovers, (d) higher crossover density, and (e) higher gamma values estimated from gamma distribution (solid bars) or modified gamma distribution (hatched bars). (f) CoC curves show that all chromosomes have the same CO interference. (g) Crossover numbers and (h) CoC curves of male mice (data provided by Lorida Anderson). Mouse autosomes are divided into three groups according to axis lengths (long chromosomes: chromosomes 1, 2, 4, 5, 7, and 11; medium chromosomes: chromosomes 3, 6, 8, 9, 10, 12, 13, and 14; short chromosomes: chromosomes 15–19). CoC analyses show that all chromosomes have the same crossover interference (curves with solid lines) as confirmed by simulations (curves with dot lines). Smax= 3.5, L = 6 μm, cL = 0.6, cR = 1, E = 0.6, B = 0.6, N is adjusted according to axis length (14, 10, and 8 for long, medium, and short chromosomes, respectively). CO: crossover; CoC: coefficient of coincidence.
