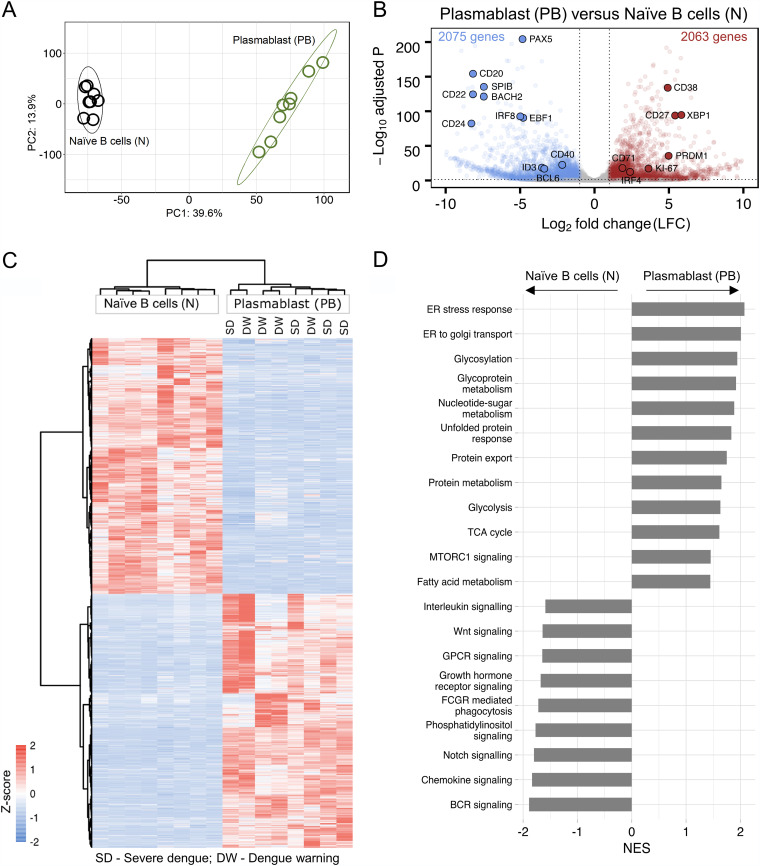
FIG 2.
Transcriptional profiling of the plasmablasts from dengue patients. (A) Principal-component analysis (PCA) score plot of normalized counts of 14,773 genes showing clustering of plasmablasts (green) and naive B cells (black) from eight different acute dengue febrile subjects. (B) Volcano plot with −log-adjusted P values on the y axis and log2 fold changes on the x axis showing 4,138 differentially expressed genes that are significantly upregulated (red dots) (right), and downregulated (blue dots) (left) in plasmablasts versus naive B cells. Gray dots are either nonsignificant, nondifferential, or both (for significant, adjusted P value of <0.05 and P value of <0.01; for differential, log2 fold change of greater than or equal to1 or less than or equal to −1). Selected genes with known expression in plasmablasts and naive B cells are labeled in the plot with larger dots and their symbol. (C) Heat map showing hierarchical clustering of 4,138 differentially expressed genes in plasmablasts compared to naive B cells from eight acute dengue febrile patients. z-scores of normalized counts were taken for the heat map. The Ward.D2 method was used for clustering (for significant, adjusted P value of <0.05 and P value of <0.01; for differential, log2 fold change greater than or equal to 1 or less than or equal to −1). The disease severity of the individual patients is indicated at the top. (D) Gene set enrichment analysis (GSEA) performed using gene sets derived from GO biological process, Hallmark, KEGG, and Reactome gene sets from MSigDB. Significant pathways with an FDR q value of <25% are shown. Enriched term names are manually collapsed. Positive and negative normalized enrichment scores (NES) correlate with upregulated and downregulated pathways, respectively, in plasmablasts compared to naive B cells. ER, endoplasmic reticulum; TCA, tricarboxylic acid; GPCR, G-protein-coupled receptor; FCGR, Fc gamma receptor; BCR, B-cell receptor.