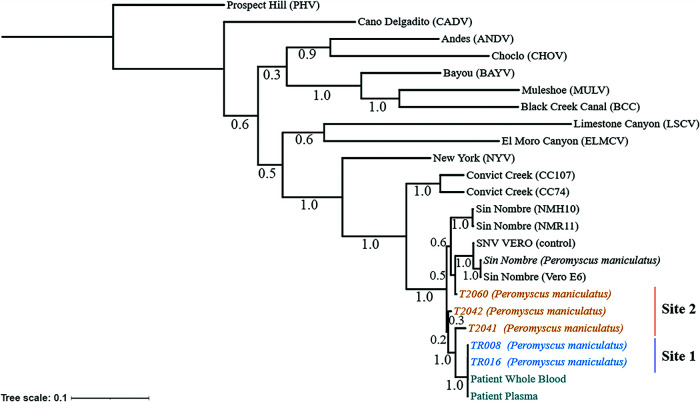
FIG 2.
Phylogenetic analysis of captured P. maniculatus and patient samples suggests transmission at the residential site (site 1). A maximum likelihood tree was generated based on concatenated S, M, and L segments. Bootstrap support is indicated. Both sites were within a 15-mile radius and are indicated as residential site (TR [site 1]) (blue) or second site (T2 [site 2]) (orange). Patient plasma and whole blood were sequenced and are shown (teal). The tree was rooted to the Prospect Hill reference sequence. Additional hantavirus reference sequences were obtained through the NCBI. SNV from infected Vero E6 cells (SNV VERO) was used as a positive sequencing control. The bar represents the number of substitutions per site.