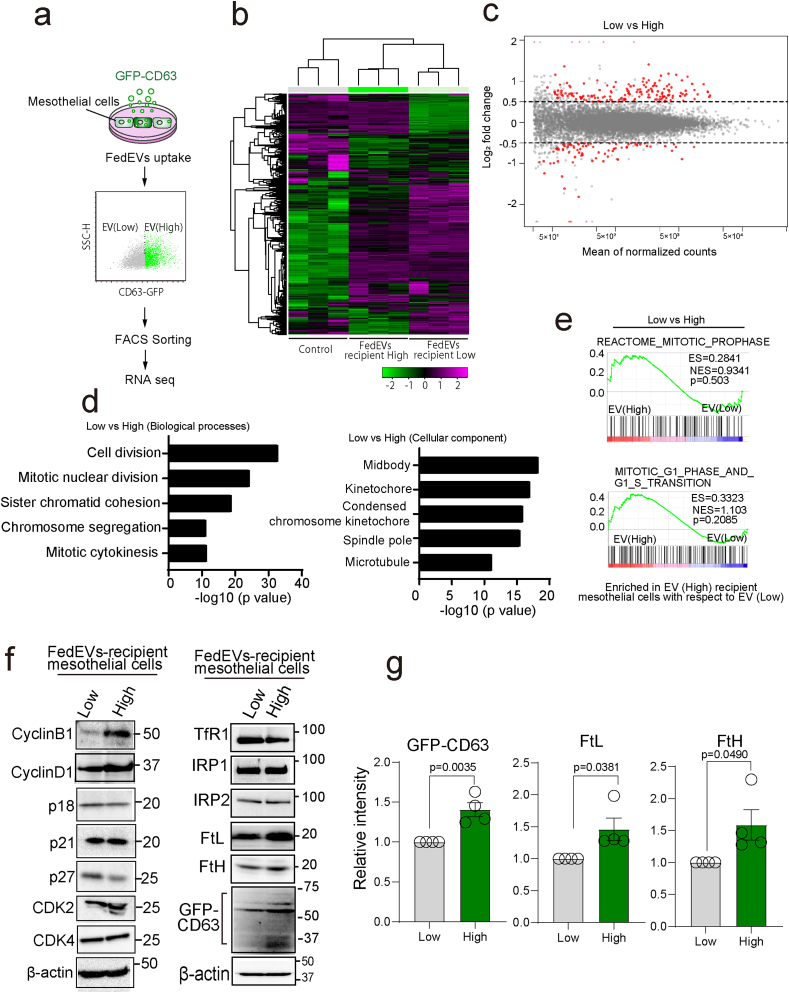
Fig. 4.
FedEVs-high mesothelial cells are under mitosis. (a) Schema of culture system and FACS sorting for RNA-sequencing analysis of MeT-5A mesothelial cells with differential phagocytosis of FedEVs. (b) Comparison of transcriptional profiles of mesothelial cells with non-treatment (NT), FedEVs low-uptake (Low) and FedEVs high-uptake (High). Magenta and green colors indicate upregulation and downregulation, respectively. (c) MA-plots of differentially expressed genes (DEGs) between low FedEVs-uptake (Low), high FedEVs-uptake (High) mesothelial cells. The X-axis represents the mean of normalized counts and the Y-axis shows the log2 fold change >0.5. Red dots correspond to statistically significant DEGs. (d) Top gene ontology (GO) biological process and cellular components terms related to DEGs in FedEVs-high MeT-5A cells. (e) Gene-set enrichment analysis revealed significant upregulation of mitosis-associated pathways with MSigDB gene set. (f) Immunoblot analysis of FedEVs-recipient mesothelial cells (Low and High). (g) Quantification of band intensity (n = 4; mean ± SEM). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)