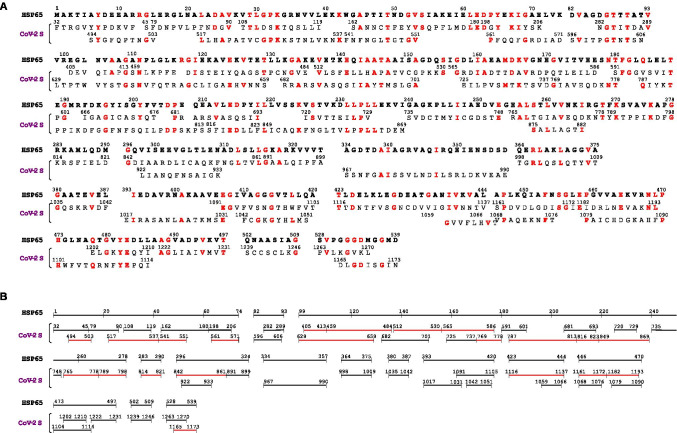
Fig. 1.
Sequence similarity between HSP65 of Mycobacterium bovis BCG and the spike (S) protein of SARS-CoV-2. Pair-wise alignment of the protein sequences was performed as specified in “Materials and methods.” A All the sequences of S protein (CoV-2 S) with similarity to HSP65, detected with both T-Coffee (upper line) and Clustal Omega (lower line), are reported below the corresponding sequences of HSP65 showing the amino acid residues (one-letter code) and sequence numbering (starting from the amino terminus of each protein). In red are the identical residues in both HSP65 and CoV-2 S. B The same sequences, as in A, are presented in graphical form to visualize the portions of CoV-2 S with similarity to HSP65. In red are the sequences identified experimentally as epitopes of CoV-2 S (see “Results”). One-letter code abbreviations for amino acid residues are the following: A, alanine; C, cysteine; D, aspartic acid; E, glutamic acid; F, phenylalanine; G, glycine; H, histidine; I, isoleucine; K, lysine; L, leucine; M, methionine; N, asparagine; P, proline; Q, glutamine; R, arginine; S, serine; T, threonine; V, valine; W, tryptophan; Y, tyrosine