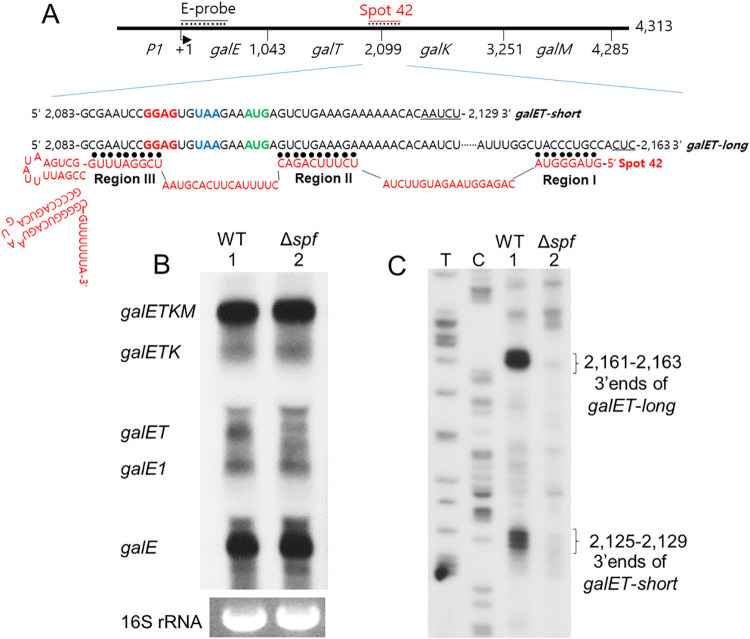
Fig 1. Spot 42 promotes generation of galET mRNA form galETKM operon.
A) Schematics of the galETKM operon showing the position of the last nucleotide of the stop codon of each gene from the transcription initiation site of the P1 promoter (+1) (top line). The Spot 42 (red) binding site spans about 75 nucleotides and is located at the galT-galK junction [20]. The region of galE used to probe northern blots is marked as E-probe. The next two lines show the nucleotide sequence of the 3’ ends of galET-short and galET-long mRNA. The letters in bold are a putative Shine-Dalgarno sequence (GGAG, red letters) for galK, the stop codon (UAA, blue letters) of galT, and the initiator codon (AUG, green letters) of galK. The sequence of the entire Spot 42 RNA is shown in red and its regions that are complementary to gal mRNA are indicated by black dots and are marked as Region I, II and III. B) Northern blot with the E probe of gal mRNA from MG1655 (WT, lane 1) and MG1655Δspf (Δspf, lane 2) cells. Note that the galET mRNA band is missing in lane 2. The identity of the two faint bands in MG1655Δspf cells where the galET band resides has not been determined. Shown at the bottom is 16S rRNA, which was used as a loading control. C) 3’ RACE assay of 3’ ends of galET mRNA from MG1655 (lane 1) and MG1655Δspf (lane 2) cells. DNA sequencing ladders that serve as length markers are in lanes marked T and C. The data presented in these blots are representative of three independent experiments.