Fig. 6.
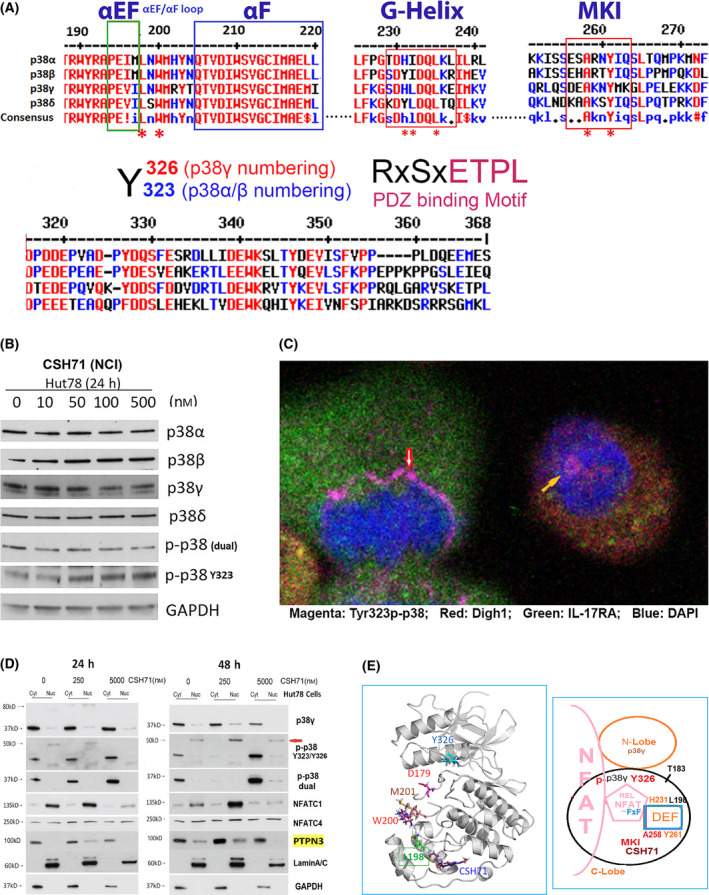
CSH71 inducing alternative p38 and NFAT activation. (A) Illustration of conserved segments of the DEF site interaction pocket (top), including α EF and α F loop, G‐Helix and MARP insert (MKI) which are structurally conserved among four p38 isoforms, which participates the formation of DEF pocket, many residues are perturbed by CSH71 binding underscored with *. The segments at bottom are p38s alternative activation site Y323/Y326 by ClustalW2 Consensus alignment diagram analysis with p38γ residue numbering. Residues ETPL is colored pink as PDZ binding Motif that solely existed in p38γ. (B) Western blot analysis to detect protein levels changes upon treatments of CSH71 (NCI) of four isoforms of p38 and two phosphorylated forms dual and alternative p38 together with GAPDH as protein internal loading control. (C) Confocal fluorescence microscopy analysis of untreated Hut78 cells indicates the subcellular location of alternative p38 activation by immunofluorescent staining using the specific antibody Y323 p‐p38. Co‐staining with antibodies against DLGH1 (red) and IL17RA (green) was used to identify downstream targets. DAPI was used to stain the nucleus. Alternative p38 activation is seen on the newly synthesized nuclear envelopes that surround anaphase chromatins (red arrow) and also within the nucleus (yellow arrow). (D) Hut 78 cell cytosolic and nuclear fractions lysates treated with indicated doses of CSH71 (250 nm and 5 µm) for 24 or 48 h are subject to WB analysis, using anti‐p38γ, anti‐p‐p38 at Y323 (alternative), anti‐p‐p38 (dual), anti‐PTPN3 and anti‐NFATC1, anti NFATC4 antibodies. Anti‐Lamin A/C antibody for the Lamin protein as the loading control for nuclear proteins and GAPDH for cytosol proteins. (E) Proposed model of interaction between NFAT with the DEF site of p38γ when the alternative site at Y326 is activated (phosphorylated). Left, 3D p38γ with residues reflect allosteric network from LBD to ATP binding site. Right, NFAT Interaction interact with p38γ via two docking motifs—FxF and LxL sequences; p38γ is also known as ERK6, which implicates similarity with the ERK family; in the REL domain of NFATc4, which contains an FxF motif (F681, F683) at the COOH‐terminal that allows interaction with the DEF pocket. Other NFAT members (NFATC1, NFATC2, and NFATC3) have FxY motifs in which possibly allows interaction with ERK. The LxL sequence is specific for other MAP kinases, such as c‐Jun NH2‐terminal protein kinase (JNK).
