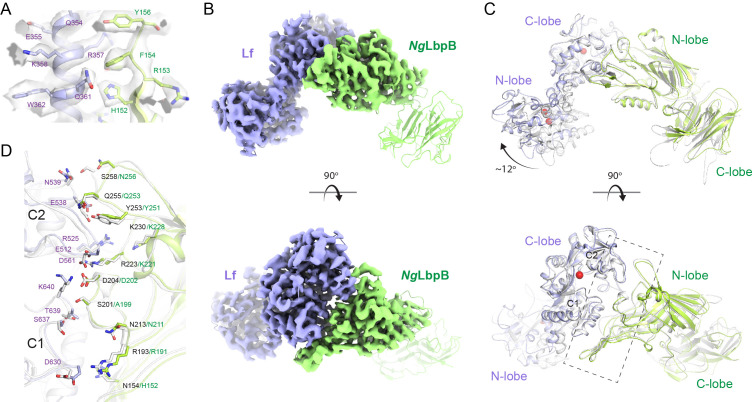
Figure 4. The 3.65 Å cryoEM structure of N. gonorrhoeae LbpB (NgLbpB) in complex with human lactoferrin.
(A) Zoomed view at the interface between NgLbpB and lactoferrin (Lf) depicting the quality of the density shown as a grey isosurface. (B) Orthogonal views of the full cryoEM map with NgLbpB in green and Lf in violet. (C) Orthogonal view of an alignment of the NgLbpB–Lf cryoEM structure (green/violet) with the N. meningitidis LbpB (NmLbpB)–Lf crystal structure (grey) (RMSD 1.4 Å along the interacting domains). (D) A zoomed view of the binding interface shows extensive interactions along an elongated surface covering both the C1 and C2 domains of Lf (buried surface area 1604 Å2).
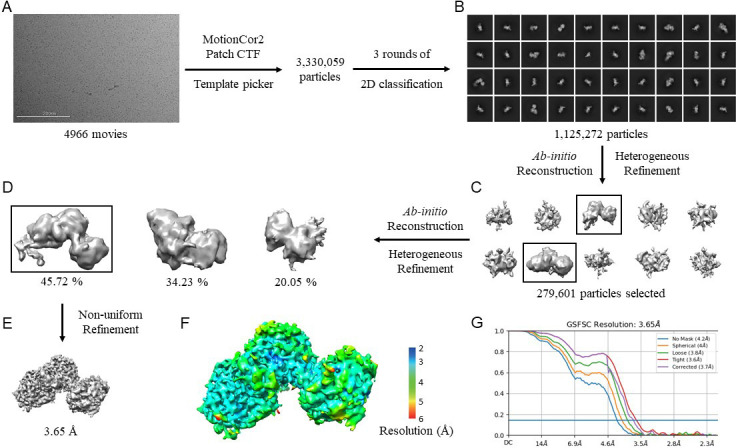
Figure 4—figure supplement 1. CryoEM data processing workflow for N. gonorrhoeae LbpB (NgLbpB)–Lf complex.