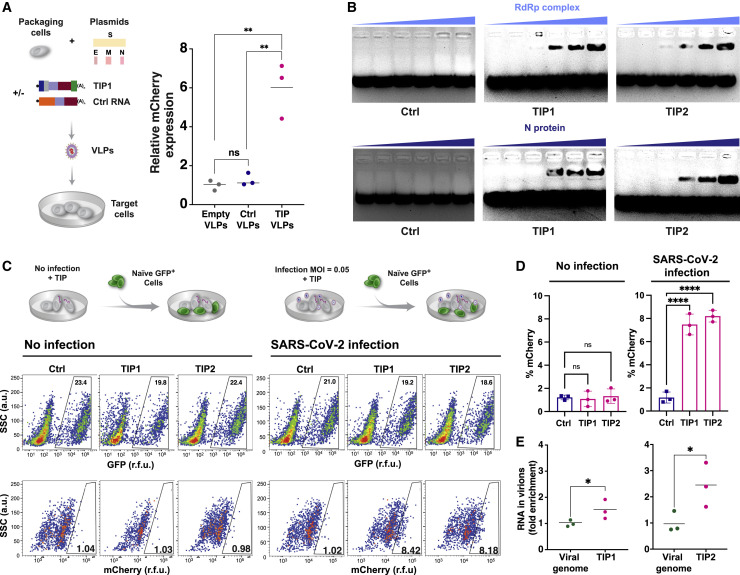
Figure 4.
TIP RNAs form functional VLPs, bind SARS-CoV-2 RdRp and N trans elements, and mobilize with R0 >1
(A) Reconstitution assay: schematic and quantification of VLP reconstitution for TIP1 and Ctrl RNA. Quantification in target cells by qRT-PCR for mCherry as compared to empty (RNA-free) VLPs.
(B) EMSA of TIP RNA or Ctrl RNA incubated with increasing concentrations of N protein or RdRp complex from cell extracts.
(C) R0 estimation via 1st-round supernatant transfer. TIP-transfected cells were infected with SARS-CoV-2 (MOI = 0.05) and then thoroughly washed to remove virus, and at 2 h post-infection GFP+ reporter cells were introduced to the culture. At 12 h post-infection, GFP+ cells were analyzed by flow cytometry to quantify the percentage mCherry+ cells (via indirect immunofluorescence staining) within the GFP+ population. Uninfected cells were used as an experimental control to confirm that TIP mobilization only occurred in the presence of SARS-CoV-2.
(D) Flow-cytometry quantification of (C).
(E) Relative packaging of TIP RNA in virions. Cells were nucleofected with TIP1 or TIP2 followed by SARS-CoV-2 infection (MOI = 0.05), and the supernatant was harvested at 24 h post-infection and analyzed by qRT-PCR for TIP RNA (using mCherry qPCR primers) versus viral genomic RNA (using E gene qPCR primers). Standard curves (see Figure S3F) were statistically indistinguishable for both primer sets.
For all panels: ns, not significant, ∗∗∗∗p < 0.0001, ∗∗p < 0.01, ∗p < 0.05 from Student’s t test. See also Figure S3.