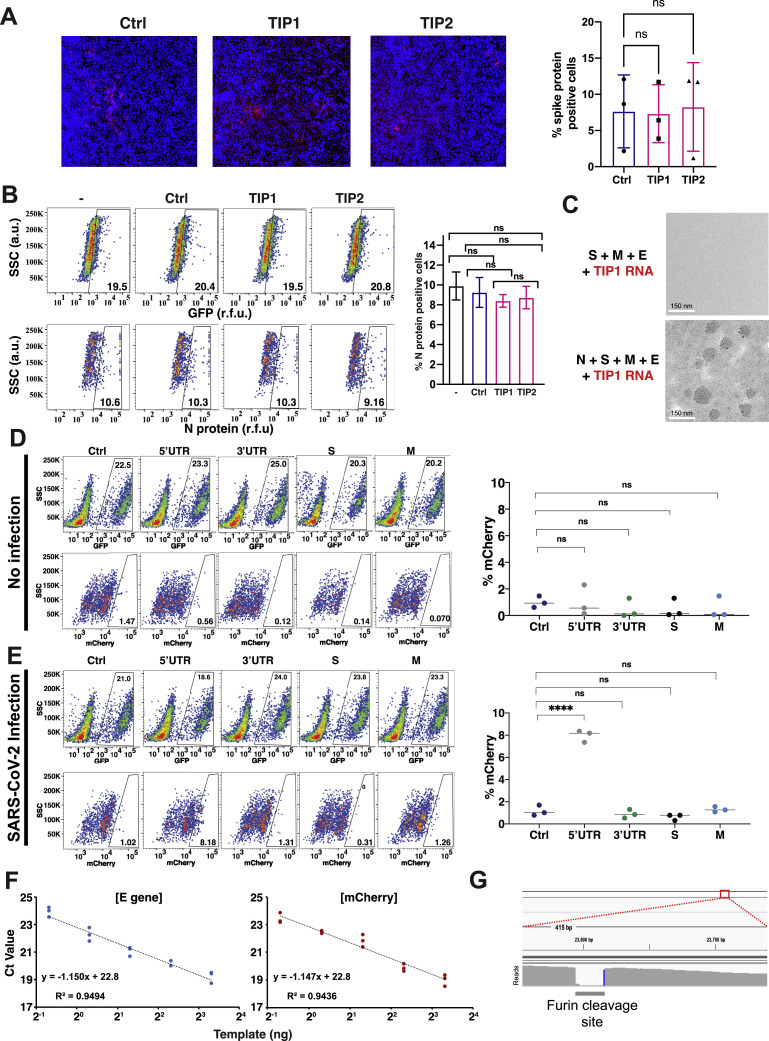
Figure S3.
Controls for TIP mechanism of action and mobilization in cell culture, related to Figure 4
(a) TIPs do not interfere with SARS-CoV-2 virus entry. Entry analysis of Vero cells nucleofected with Ctrl, TIP1 or TIP2 RNAs. 24 hours post nucleofection, cells were infected with SARS-CoV-2 (WA-1 isolate; MOI = 20). Two hours post infection, cells were fixed, stained for S protein and nucleus (DAPI), and subjected to high-throughput confocal microscopy. Left: representative images for overlay of S and nucleus. Right: quantification of % S protein positive cells in all three samples. ns, not significant. (b) TIPs do not interfere with early events in SARS-CoV-2 life cycle. Flow cytometry of Vero cells nucleofected with Ctrl, TIP1 or TIP2 RNAs and infected (MOI = 0.05) at 24 hours post nucleofection. Two hours post-infection cells were thoroughly washed and naive GFP+ cells were added. Cells were harvested at 8 hours post infection (∼one replication cycle), stained for N protein and subjected to flow cytometry. Left: flow cytometry dot plots; Right: bar graph representing % N protein positive cells. ns, not significant. (c) Transmission EM analysis of concentrated supernatant from cells transfected with S, E, and M and TIP RNA showing no VLP (top) and from concentrated supernatant from cells transfected with N, S, E, and M and TIP RNA showing VLPs (bottom), Scale bar = 150nm. (d, e) R0 estimation via 1st round supernatant transfer for Ctrl RNA, 5′UTR, 3′ UTR, S(3xTAA) and M(3xTAA) controls in the absence (d) and presence (e) of infection (MOI = 0.05). TIP-transfected cells were infected with SARS-CoV-2 or mock, thoroughly washed, and at two hours post-infection or mock infection, GFP+ reporter cells were introduced to the culture. At 12 hours post infection, GFP+ cells were analyzed by flow cytometry to quantify % mCherry+ cells (by indirect immunofluorescence) within the GFP+ population. (f) Standard curve of E-gene and mCherry using qPCR at the mentioned concentrations. (g) Alignment of sequencing from long-term culture showing furin cleavage site mutation. (For all panels: Error bars represent standard deviation from three biological replicates; ns denotes not significant, ∗∗∗∗ denotes p < 0.0001, Student’s t test.)