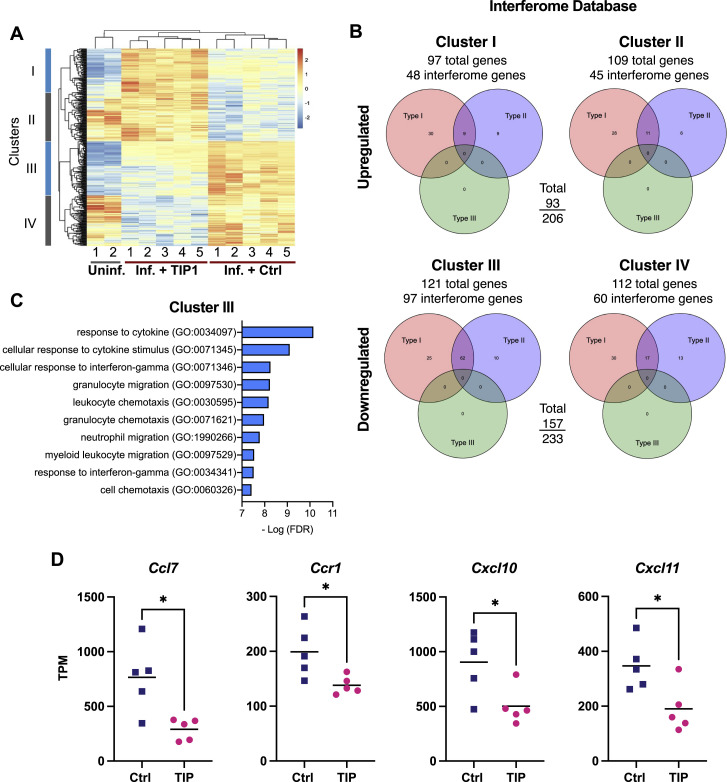
Figure S5.
TIP-treatment mitigates overexpression of pro-inflammatory genes in SARS-CoV-2-infected hamster lungs, related to Figure 7
(a) Differential gene expression in hamster lungs on day 5 post infection by RNaseq analysis. Each column represents one animal clustered by expression profiles. Uninfected hamster data was obtained from GSE157058 (Sahoo et al., 2021). DEGs were defined by comparing infected samples treated with TIP RNA or Ctrl RNA LNPs, and are grouped in four clusters. (b) Venn diagram of RNA sequencing of hamster lungs summarizing differentially expressed genes (DEGs) in TIP versus Ctrl-treated animals using the Interferome database http://www.interferome.org with parameters Mus musculus to approximate Syrian golden hamster. According to the Interferome database, the majority of the DEGs in cluster III are interferon-stimulated genes (ISGs), regulated by either Type I or Type II interferons (IFNs). (c) Gene ontology (GO) analysis showing the top ten biological processes enriched in cluster III. (d) Expression levels in terms of transcripts per million (TPM) for additional representative genes belonging to cytokine/chemokine pathways (individual animals are shown as individual data points). These proinflammatory cytokines (Ccl7, Ccr1, Cxcl10, Cxcl11) were previously reported to be upregulated in COVID-19 patients but are significantly reduced in TIP-treated animals. [For all panels: ∗ denotes p < 0.05 from Student’s t test].