Figure 2.
PI3P production is required for efficient HCV and SARS-CoV-2 replication
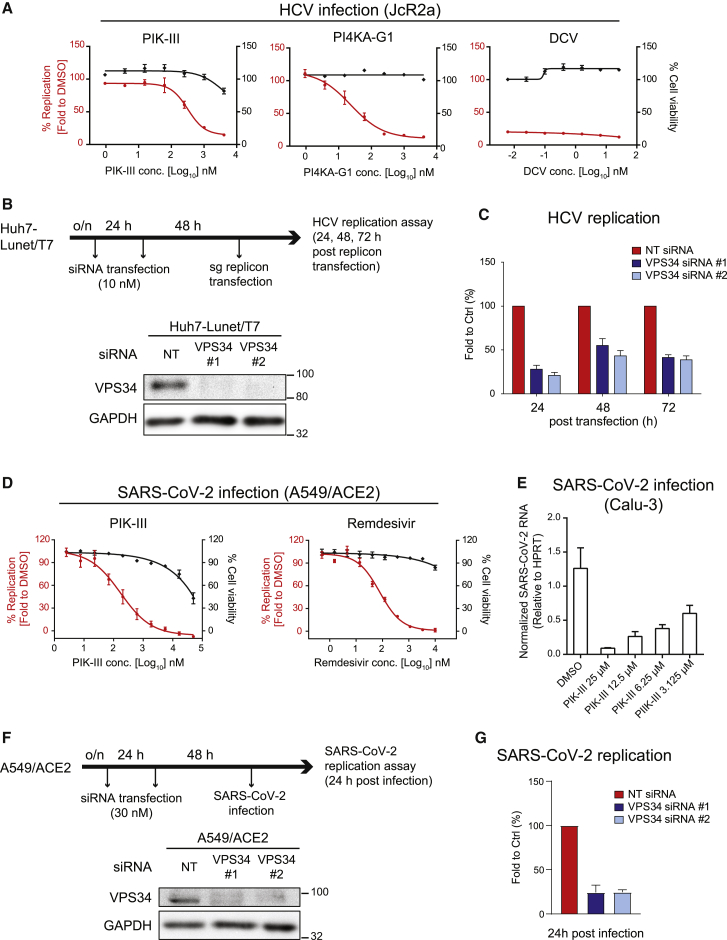
(A) Huh7-Lunet/CD81H cells were infected with the HCV reporter virus JcR2a and treated with given drugs 4 h after infection. Luciferase activity reflecting viral RNA replication (red lines) and cell viability (ATP content; black lines) was measured at 24 h post-infection. DCV, daclatasvir.
(B) (Upper) Schematic depiction of the experimental setup. Huh7-Lunet/T7 cells were transfected twice with two different VPS34-targeting siRNAs using a 24-h interval, prior to electroporation with a subgenomic HCV reporter replicon 48 h after the second siRNA transfection. (Lower) knockdown (KD) efficiency was determined by western blotting. GAPDH served as loading control.
(C) Luciferase activity reflecting HCV RNA replication was measured 24, 48, and 72 h after electroporation. For each replicon RNA transfection, values were normalized to the 4-h value reflecting transfection efficiency, and these values were normalized to those obtained with NT siRNA-transfected cells. Data in (A) and (C) represent the mean ± SEM from three independent experiments.
(D) A549/ACE2 cells were treated with serial dilutions of given drugs 30 min before infection with SARS-CoV-2 (MOI of 1). Infected cells were fixed 24 h post-infection and the percentage of inhibition was determined using N protein-specific immunostaining. Cell viability (ATP content) was measured at the same time point.
(E) Calu-3 cells were treated with PIK-III 2 h after infection with SARS-CoV-2 (MOI of 5). Total RNA of infected cells was extracted at 6 h post-infection and analyzed using qRT-PCR to determine replication efficiency.
Data in (D) and (E) represent the mean ± SEM from two independent experiments.
(F) (Upper) Schematic of the experimental setup. A549/ACE2 cells were transfected twice with siRNAs using a 24-h interval, prior to SARS-CoV-2 infection 48 h after the second siRNA transfection. (Lower) Confirmation of KD efficiency by western blotting. GAPDH served as loading control.
(G) Percentage of SARS-CoV-2 replication was determined using N protein-specific immunostaining at 24 h post-infection. Data represent the mean ± SEM from three independent experiments. See also Figure S2.