Fig. 4.
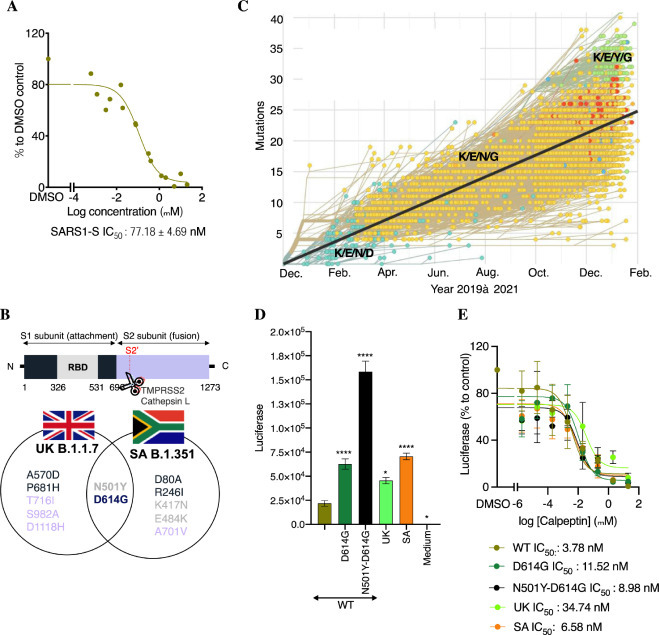
Breath of activity of calpeptin against various SARS-CoVs. A. Its activity against SARS1-S in HEK293T-ACE2 cells. HEK293T-ACE2 cells were incubated with different concentrations of calpeptin, then infected with SARS1-S. Luciferase was measured 48 h later, using Bright-Glo. Shown is the mean ± SEM of n = 2 independent experiments. B. Schematic of the substituted residues in the S protein of the highest threat of SARS-CoV-2 strains. C. Evolution of the S protein residues at the position 417, 484, 501 and 614 from 2019 to February 2021. Modified figure from https://nextstrain. org/ncov/global?branchLabel=none&c=gt-S_417,484,501,614&l =clock. D. Activity of the new emergent variants. HEK293T-ACE2 cells were infected with different mutants of SARS2-S. The day after, a medium change was performed. Luciferase was measured 48 h later, using Bright-Glo. Shown is the mean ± SEM of n = 3 independent experiments. WT: wild type, SA: South Africa, UK: United Kingdom. E. Activity of calpeptin activity against crucial mutations present in the S protein of the new emergent strains. Similar experiment than D but calpeptin was added during infection and after medium change. Shown is the mean ± SEM of n = 2–5 independent experiments. Two-way ANOVA followed by Dunnett's post-test were used for statistical comparisons. *, P < 0.01; **, P< 0.001; ***, P < 0.0001.
