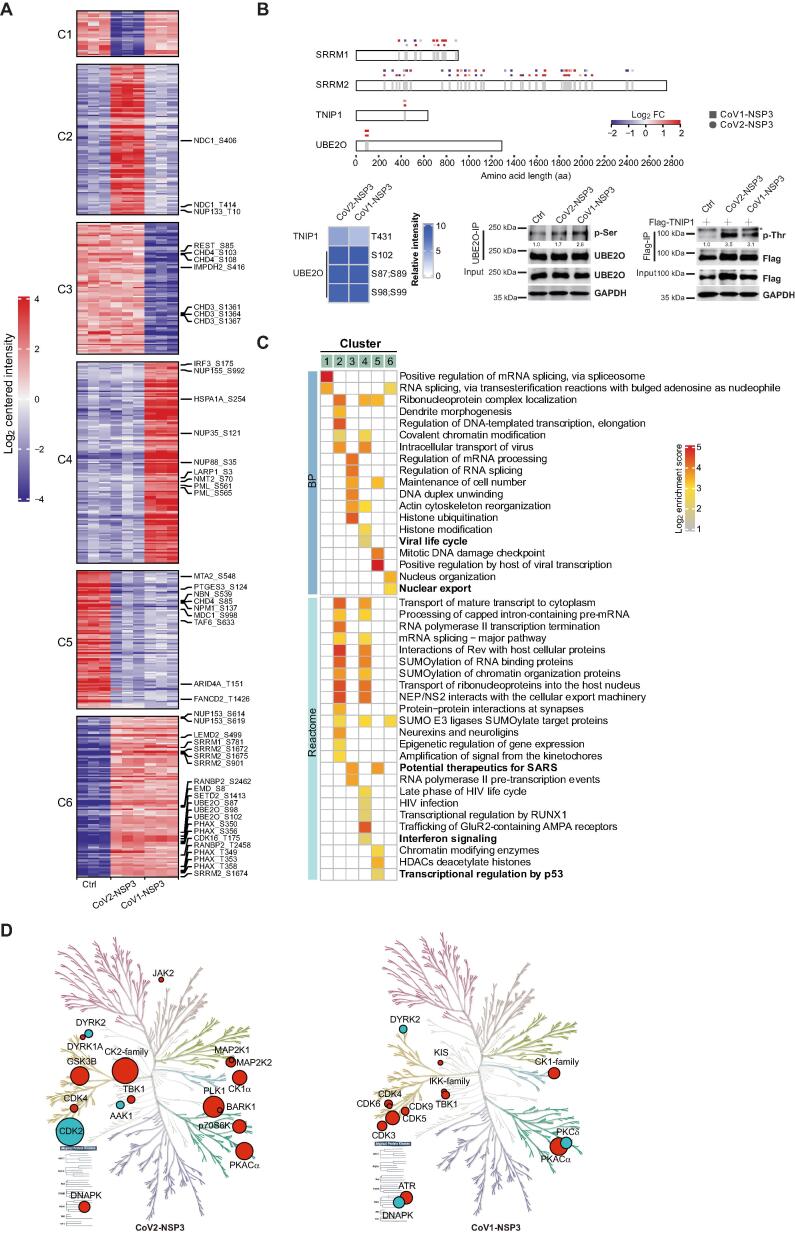
Figure 5.
Phosphoproteome analysis of NSP3-expressing cells
A. Clustering of DEPPs in NSP3-expressing cells. DEPPs showing significantly differences in CoV1-NSP3- and CoV2-NSP3-expressing cells compared to control cells were clustered together. C1–C6 on the left indicate Clusters 1–6. Full list of DEPPs is available in Table S5. Representative DEPPs with highlighted GO in (C) were shown on the right. B. Validation of phosphoproteome studies. Top panel: schematic representation of phosphorylated sites of indicated proteins. Square indicates a DEPP with significant change in CoV1-NSP3-expressing cells, whereas circle indicates a DEPP with significant change in CoV2-NSP3-expressing cells. Bottom panels: HEK293T cells transfected with indicated plasmids were subjected to IP assays (anti-Flag-resin or anti-UBE2O antibody with protein A/G), following immunoblotting with phosphorylated serine/threonine antibodies. ImageJ was used to quantify the relative expression of each protein to its matching loading control. C. Representative enriched GO BP and Reactome terms. Labels on top of the heatmap correspond to the clusters in (A). Full list of the GO and Reactome terms is shown in Table S5. Bold terms containing DEPPs that are highlighted on the right of (A). D. KinMaps showing altered kinases whose activities mapped to DEPPs in NSP3-expressing cells. Significantly enriched DEPPs in CoV1-NSP3- and CoV2-NSP3-expressing cells were matched to kinases. Kinome profiling was performed using KinMap interface. Red indicates up-regulation, whereas cyan indicates down-regulation. Size of the circles reflects the number of DEPPs. DEPP, differentially enriched phosphorylated peptide.